The Impact of Animal-Free Antibodies on the Future of Antibodies
1. Introduction to Antibodies
2. Antibody Production
3. Applications of Antibodies in Science
3.1 Basic Research
3.2 Therapeutics
4. Overcoming Immunogenicity
4.1 Chimerization
4.2 Humanization
4.3 Fully Human
5. Advantages of Animal-Free Antibodies (AFAs)
6. Next Generation Antibody Production
6.1 Artificial Intelligence
6.2 Bispecific Antibodies
7. Conclusion
8. Providers
9. Abbreviations
10.References
2. Antibody Production
3. Applications of Antibodies in Science
3.1 Basic Research
3.2 Therapeutics
4. Overcoming Immunogenicity
4.1 Chimerization
4.2 Humanization
4.3 Fully Human
5. Advantages of Animal-Free Antibodies (AFAs)
6. Next Generation Antibody Production
6.1 Artificial Intelligence
6.2 Bispecific Antibodies
7. Conclusion
8. Providers
9. Abbreviations
10.References
1. Introduction to Antibodies
In the 1890s, antibodies were first detected in blood plasma where they neutralized diphtheria toxin. In 1939 immunized serum was separated by electrophoresis and later by weight using a chromatography column, giving the first indication of multiple antibody subtypes 1. Their structure was only fully discovered in the 1960s. Antibodies, also called immunoglobulins (Ig), are created from two light (L) and two heavy (H) chains connected by disulfide bonds 2. These chains consist of a combination of variable (V) and constant (C) regions. L chains have one of each and H chains have one V region and three Cs. Whole antibodies consist of different fragments that can be isolated through genetic or biochemical means for antibody engineering 3
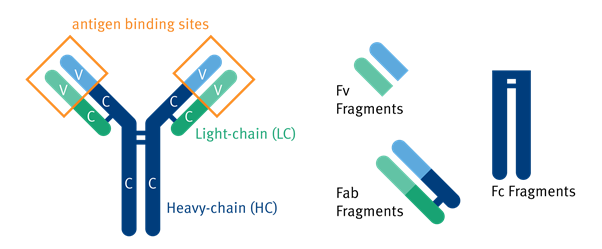
Figure 1.1: Illustration of the components of the human antibody including Fv, Fab, and Fc Fragments 4.
H chain variability is responsible for the five different human antibody types; IgG, IgM, IgD, IgE, and IgA 7. IgG is the most abundant in the body making up to 70-75% 8. Each immunoglobulin has its own specific function:
IgM is the neutralizing antibody against bacterial toxins 9. It tags the antigen for death by complement fixation, in which the classical pathway is triggered causing the membrane attack complex to activate. It is part of the primary immune response and makes up the ‘natural’ antibodies as they are generated early in B cell response; they have a lower affinity for specific antigens but can bind more broadly to pathogens 10.
IgG has 4 subtypes: IgG1, IgG2, IgG3, and IgG4. IgG1 and IgG3 work against proteins, and IgG2 and IgG4 against bacterial polysaccharides 1, 9. They too activate the complement pathway and play a role in the neutralization of pathogens. IgG3 has been the most effective against human immunodeficiency virus (HIV) 9. IgG4 is unable to fix complement and the other 3 differ in their binding affinities for component 1q (C1q) of the complement pathway. All 4 subtypes also take part in the secondary immune response. IgG4 is inhibitory in autoimmune diseases as it blocks the activation of the complement system by IgG1 and IgG3 autoantibodies 11.
Autoantibodies are antibodies that react with self-antigens. The unproblematic autoantibodies are generally IgM, due to their production early in B cell development leading to unmutated genes that have a slight affinity for their own antigens. These autoantibodies play a role in the homeostasis of the immune system. Problems occur when autoantibodies are formed from mutated IgG molecules. These disrupt cell signaling and clearance. They can arise before the onset of autoimmune disorders so have potential as a diagnostic biomarker 12.
IgD through class-switching from IgM in the respiratory tract stimulates B-cells to secrete more IgD. This causes activation of the mucosal immunity; IgD binds both microbial virulence factors and pathogens, initiating the immune response against respiratory pathogens. The secretion of IgD-producing B-cells into the circulation causes cross-linking of basophils, arming them with immune cell mediators as well as activating other immune cells 13. It has been hypothesized that IgD can control B-cell fate by changing the activation status at developmental stages 1. IgD does not initiate the complement pathway and participation in other pathways is poorly understood as of yet.
IgA has 2 subtypes; IgA1 and IgA2. They make up almost 50% of the protein in colostrum which is thought to be crucial for newborns’ passive immune system. IgA1 is more prominent in serum whereas IgA2 is more prominent in mucosa due to better resistance to proteases 1. Their predominant sites indicate post-translational function. IgA2 is involved in protection against mucosal infection and IgA1 is involved in triggering effector functions to destroy microorganisms. IgA does not play a role in the complement pathway but recently it has been noted that when bound to mannan-binding lectin can initiate the lectin pathway 14. IgA receptors are present on neutrophils indicating that they also induce the local antibody-dependent cellular cytotoxic (ADCC) response 1.
IgE is very potent such that it causes upregulation of FcεR and has a very high binding affinity. It is associated mainly with allergic reactions. This allergic response, caused by the cross-linking of FcεRI in response to IgE on mast cells, basophils, eosinophils, and Langerhans cell degranulation, presents itself as an inflammatory response 1. This inflammatory response can be as small as a sniffle or as severe as anaphylaxis 15. An antibody to counteract this response has been devised, it binds to free IgE preventing this binding event. IgE also plays a key role in the protection against parasites. It does not participate in the initiation of the complement pathway but protects by initiating cascades that cause vasodilation. These functions have also seen the hypothesized use of IgE as an anticancer agent 16.
IgM is the neutralizing antibody against bacterial toxins 9. It tags the antigen for death by complement fixation, in which the classical pathway is triggered causing the membrane attack complex to activate. It is part of the primary immune response and makes up the ‘natural’ antibodies as they are generated early in B cell response; they have a lower affinity for specific antigens but can bind more broadly to pathogens 10.
IgG has 4 subtypes: IgG1, IgG2, IgG3, and IgG4. IgG1 and IgG3 work against proteins, and IgG2 and IgG4 against bacterial polysaccharides 1, 9. They too activate the complement pathway and play a role in the neutralization of pathogens. IgG3 has been the most effective against human immunodeficiency virus (HIV) 9. IgG4 is unable to fix complement and the other 3 differ in their binding affinities for component 1q (C1q) of the complement pathway. All 4 subtypes also take part in the secondary immune response. IgG4 is inhibitory in autoimmune diseases as it blocks the activation of the complement system by IgG1 and IgG3 autoantibodies 11.
Autoantibodies are antibodies that react with self-antigens. The unproblematic autoantibodies are generally IgM, due to their production early in B cell development leading to unmutated genes that have a slight affinity for their own antigens. These autoantibodies play a role in the homeostasis of the immune system. Problems occur when autoantibodies are formed from mutated IgG molecules. These disrupt cell signaling and clearance. They can arise before the onset of autoimmune disorders so have potential as a diagnostic biomarker 12.
IgD through class-switching from IgM in the respiratory tract stimulates B-cells to secrete more IgD. This causes activation of the mucosal immunity; IgD binds both microbial virulence factors and pathogens, initiating the immune response against respiratory pathogens. The secretion of IgD-producing B-cells into the circulation causes cross-linking of basophils, arming them with immune cell mediators as well as activating other immune cells 13. It has been hypothesized that IgD can control B-cell fate by changing the activation status at developmental stages 1. IgD does not initiate the complement pathway and participation in other pathways is poorly understood as of yet.
IgA has 2 subtypes; IgA1 and IgA2. They make up almost 50% of the protein in colostrum which is thought to be crucial for newborns’ passive immune system. IgA1 is more prominent in serum whereas IgA2 is more prominent in mucosa due to better resistance to proteases 1. Their predominant sites indicate post-translational function. IgA2 is involved in protection against mucosal infection and IgA1 is involved in triggering effector functions to destroy microorganisms. IgA does not play a role in the complement pathway but recently it has been noted that when bound to mannan-binding lectin can initiate the lectin pathway 14. IgA receptors are present on neutrophils indicating that they also induce the local antibody-dependent cellular cytotoxic (ADCC) response 1.
IgE is very potent such that it causes upregulation of FcεR and has a very high binding affinity. It is associated mainly with allergic reactions. This allergic response, caused by the cross-linking of FcεRI in response to IgE on mast cells, basophils, eosinophils, and Langerhans cell degranulation, presents itself as an inflammatory response 1. This inflammatory response can be as small as a sniffle or as severe as anaphylaxis 15. An antibody to counteract this response has been devised, it binds to free IgE preventing this binding event. IgE also plays a key role in the protection against parasites. It does not participate in the initiation of the complement pathway but protects by initiating cascades that cause vasodilation. These functions have also seen the hypothesized use of IgE as an anticancer agent 16.
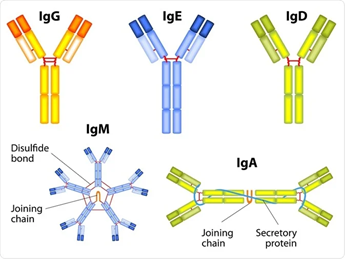
Figure 1.2: Visual representation of the 5 subtypes of immunoglobulins, including the hypothesized shape 17.
Antibodies are secreted into circulating blood and other bodily fluids by B cells in response to an antigen. This is part of the adaptive immune response. In circulation they are responsible for antigen binding, preventing the antigen from binding to cellular receptors causing initiation of infection. Instead of successfully infiltrating the cell, antigens are tagged for destruction by the immune system and are engulfed by phagocytes 18. When a B cell secretes an antigen it is specific to only one epitope on the antigen, these are known as monoclonal antibodies (mAbs). Polyclonal antibodies (pAbs) are created from the culmination of multiple antibodies produced by the binding of individual B cells to unique epitopes on the antigen or multiple different antigens. These have a better affinity with the antigen as they can target multiple epitopes at a time 19, 20. Natural antibodies are monospecific, so only bind to one target or one epitope but can be bivalent if the antigen-binding regions are the correct distance apart 21. Valency is also dependent on how many antigen-binding sites the antibodies contain. Bivalent antibodies have two binding sites 21, one for each antigen, tri has three, tetra has four, and so on 22.
MAbs are more generally used in research and therapeutics and as such have a nomenclature of their own in the form of international non-proprietary names (INN). The name gives us a full picture of the antibody, how it was produced, what system it was created with etc. If the name ends in -pab or -mab then it is a polyclonal or monoclonal antibody (pab is rarely used). The 1 or 2 letters before the -mab are indicative of the type of antibody, -ximab is chimeric, -omab is murine, – zumab is humanized, and -umab is fully human. The collection of letters before this indicates the target molecule for this antibody, such as pertuzumab, for example, is a humanized mAb that targets the tumor cells (-tuzumab). A more extensive list of targets and sources/types can be seen in Table 1.1 23, 24, 25.
MAbs are more generally used in research and therapeutics and as such have a nomenclature of their own in the form of international non-proprietary names (INN). The name gives us a full picture of the antibody, how it was produced, what system it was created with etc. If the name ends in -pab or -mab then it is a polyclonal or monoclonal antibody (pab is rarely used). The 1 or 2 letters before the -mab are indicative of the type of antibody, -ximab is chimeric, -omab is murine, – zumab is humanized, and -umab is fully human. The collection of letters before this indicates the target molecule for this antibody, such as pertuzumab, for example, is a humanized mAb that targets the tumor cells (-tuzumab). A more extensive list of targets and sources/types can be seen in Table 1.1 23, 24, 25.
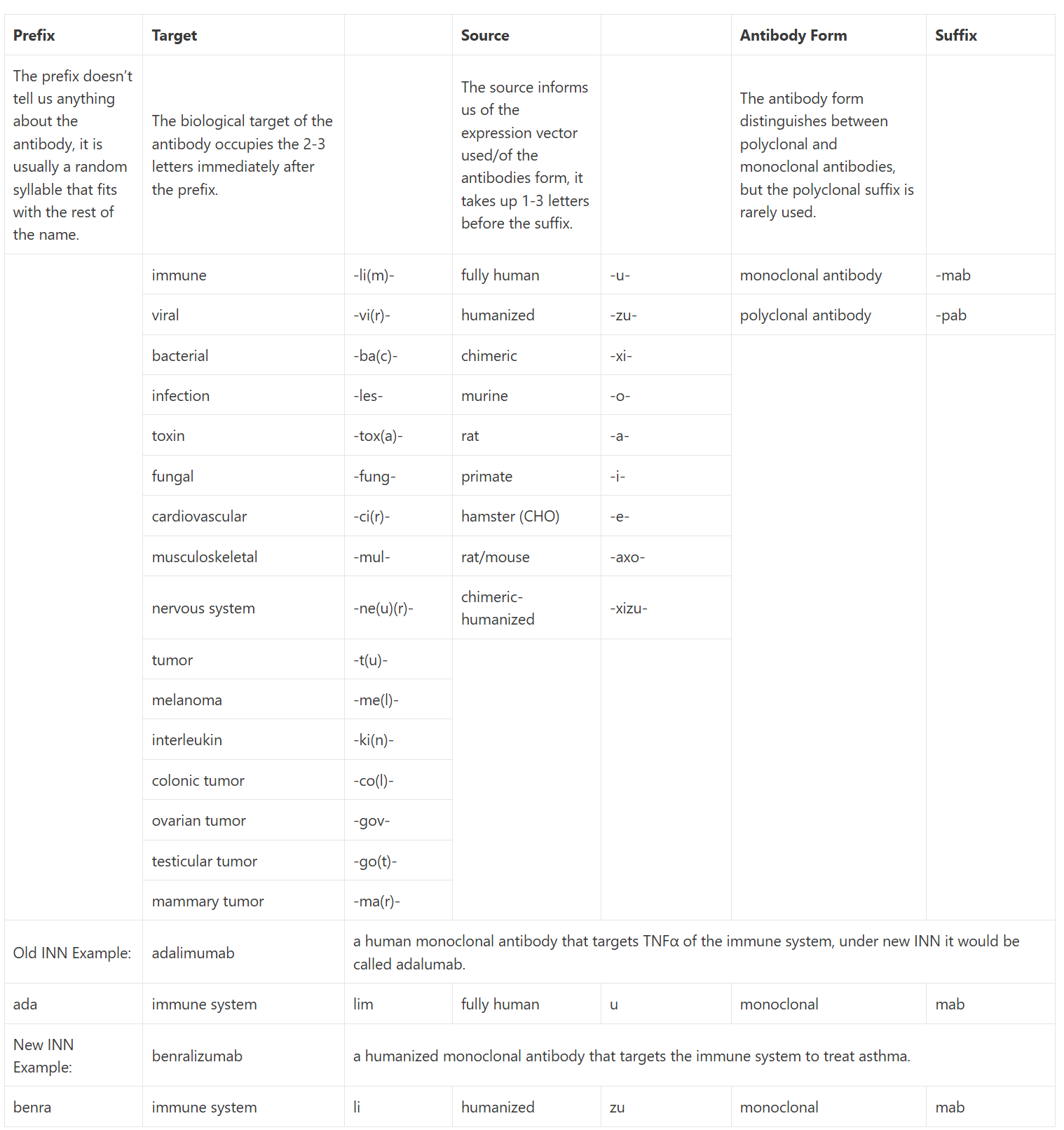
Table 1.1: This table includes both the old international nonproprietary names (INN), pre-2014 in orange, and the new naming convention, post-2017 in black. The more specific tumor nomenclature has been replaced with the generalized -tu- as antibodies can target more than one type at a time. Within the new nomenclature, many of the target shorthand can be shortened further, the last letter may be omitted to make the name easier to pronounce, adalimumab is a perfect example of this; under the new nomenclature, it would be adalumab as adaliumab would be too hard to say. Many of the antibodies approved before 2014 still go by the old nomenclature so it is useful to have both to hand. This table was created from a culmination of information from Immunology For Pharmacy 23, Concepts in Immunology and Immunotherapeutics 24, and Bioatla’s Educational Appendix 25.
2 Antibody Production
Both pAbs and mAbs have traditionally been produced with the use of animals. This involves injecting an antigen with an adjuvant into an animal, usually mice or rabbits, but the choice depends on the quantity of antibody to be produced. This triggers the immune response and the antiserum is then collected and purified from the blood 26. This is where pAb production stops.
For mAbs, the process has some additional steps. Instead of collecting the antiserum, the B cells from the spleen are collected and fused to myeloma cells to produce a hybridoma that secretes monospecific antibodies 20. This can be performed in vitro or in vivo through the ascites method but is very cruel as it involves the killing of at least two mice if not more. The first mouse is immunized and killed for its B cells, the second is injected with an inflammatory agent and the hybridoma causing a distended stomach from which the antiserum is extracted 2-3 times before the mouse dies and more are required for testing and validation of these antibodies 26. The in vitro method of hybridoma generation, while only requiring the use of one mouse for B cell extraction, is no better where animal welfare is concerned 26. Immune responses can be severe enough for the animals to show visible pain such as hunching over 27. In response to this cruelty the German, UK, Netherlands, and Australian governments have moved to ban the ascites method with the US also encouraging alternative methods 28, 29.
Animal-derived antibodies take months to generate; pAbs can take anywhere from 2-4 months and mAbs take approximately 6 months. pAbs are also susceptible to batch variation and low-specificity due to the inclusion of multiple antibodies. Both are also prone to high background and undefined identity of the binding reagent 30.
Recombinant antibodies have been around since the 1990s and are more widely adopted recently due to the many issues associated with animal-derived antibodies. They are useful for either therapeutic or research purposes. These antibodies can be established in two ways. The first is also through immunization of animals; IgG is isolated from the B cells, cloned into a vector to be infected into cell lines producing the antibody product (the transgenic method as detailed below).
In the 2nd, the immunization step can be replaced through the use of naïve libraries of immunoglobulin encoding genes from human B cells (the display methods discussed below). Although recombinant antibodies are currently more expensive to produce, the process is quicker (weeks instead of months) 31 and can rule out the use of animals in their production, overcoming the limits of animal-derived antibodies and bringing many benefits as described later in this report 27.
For mAbs, the process has some additional steps. Instead of collecting the antiserum, the B cells from the spleen are collected and fused to myeloma cells to produce a hybridoma that secretes monospecific antibodies 20. This can be performed in vitro or in vivo through the ascites method but is very cruel as it involves the killing of at least two mice if not more. The first mouse is immunized and killed for its B cells, the second is injected with an inflammatory agent and the hybridoma causing a distended stomach from which the antiserum is extracted 2-3 times before the mouse dies and more are required for testing and validation of these antibodies 26. The in vitro method of hybridoma generation, while only requiring the use of one mouse for B cell extraction, is no better where animal welfare is concerned 26. Immune responses can be severe enough for the animals to show visible pain such as hunching over 27. In response to this cruelty the German, UK, Netherlands, and Australian governments have moved to ban the ascites method with the US also encouraging alternative methods 28, 29.
Animal-derived antibodies take months to generate; pAbs can take anywhere from 2-4 months and mAbs take approximately 6 months. pAbs are also susceptible to batch variation and low-specificity due to the inclusion of multiple antibodies. Both are also prone to high background and undefined identity of the binding reagent 30.
Recombinant antibodies have been around since the 1990s and are more widely adopted recently due to the many issues associated with animal-derived antibodies. They are useful for either therapeutic or research purposes. These antibodies can be established in two ways. The first is also through immunization of animals; IgG is isolated from the B cells, cloned into a vector to be infected into cell lines producing the antibody product (the transgenic method as detailed below).
In the 2nd, the immunization step can be replaced through the use of naïve libraries of immunoglobulin encoding genes from human B cells (the display methods discussed below). Although recombinant antibodies are currently more expensive to produce, the process is quicker (weeks instead of months) 31 and can rule out the use of animals in their production, overcoming the limits of animal-derived antibodies and bringing many benefits as described later in this report 27.
3 Applications of Antibodies in Science
3.1 Basic Research
Antibody potential applications were immediately apparent after discovery. They have made great strides in basic research with their broad-scale use to detect antigens being adapted for different testing methods. The first being their use in immunofluorescence in 1941, which enabled the localization and detection of proteins in biological specimens through the use of fluorophore-tagged antibodies 32. From there they have been included in immunohistochemical (IHC) staining since 1942 33, in enzyme-linked immunosorbent assay (ELISA) (competitive, sandwich, and indirect) established between 1971-1978 34 which enables the quantification of antibodies 35, in western blotting since 1979 36 for separation and identification of individual proteins from mixtures 37, and in flow cytometry which was discovered between 1970-1980 38 that utilizes lasers to analyze cell populations in solution 39.
These testing techniques have enabled vast amounts of research using antibodies. In studies of neurobiology, autoantibodies have been noted to play a role in autoimmune neurological diseases particularly encephalitis. With the use of recombinant patient-derived mAbs, they determined how the autoantibodies work. They cross-link proteins on the synaptic cell surface such as neurotransmitters like N-methyl-D-aspartate (NMDA) receptors and cause internalization of these receptors affecting amino-terminal domain (ATD)-mediated gated control presenting as symptoms of epilepsy, psychosis, etc. IHC staining assisted this study by providing information about the localization of these autoantibodies and the Leucine-rich glioma-inactivated 1 (LGI1) protein was noted to affect protein-protein interactions. Autoantibody potential in basic neuroscience was discovered with further uses in structure-function relationships, discovery of protein-specific roles in vivo from induced recombinant antibody (rAb) expression, epitope mapping, and analysis of protein distribution as well as studies of its own role in the body 40.
Antibodies are notoriously beneficial in proteomic studies. Their ability to bind proteins with specific characteristics provides more insight into certain diseases such as autophagy. Autophagy is caused by phosphorylation of the Autophagy Related 16 Like 1 (ATG16L1) protein in response to cellular and metabolic stresses. Through a collaboration with Abcam, the researchers designed a mAb that focuses on ATG16L1 phosphorylation on serine 278 of newly formed autophagosomes in mammalian cells. It’s important that the autophagosomes are new as dysfunctional ones can skew experimental results. Their studies with this antibody using ATG16L1 knockout and wildtype versions suggest that the rate of autophagy is controlled by another regulatory event in an uncharacterized region. Along with starvation which increases the number of new autophagosomes formed, and other autophagy inducing stresses. The success of this antibody has seen it become commercially available to all researchers and is compatible with IHC, IF, and western blotting 41.
Antibodies are known for the immunity they afford us, much of this is passed on from mother to child via colostrum and breast milk. Different factors can have separate effects on the concentrations of Igs in colostrum. One study found that malnourishment results in only a third of normal IgG concentrations and identified reductions in IgA and C4 of the complement pathway of the immune response 42. Another such study looked at the effects of immunization and infection during pregnancy on IgA colostrum concentrations. Through the use of ELISA and western blot they measured the IgA subclasses; ELISA quantified concentrations of IgA in relation to the vaccine antigens, and western blot evaluated the ability of colostrum to recognize antigens of E.coli. IgA2 was more reactive against lipopolysaccharide (LPS) and increased in women with multiple gastrointestinal (GI) infections, IgA1 increased in response to multiple doses of tetanus vaccine. The ELISA analysis also identified an increase of IgA2 in relation to the Pneumococcal polysaccharides (PPS) vaccine. Previous studies suggested that IgA-producing cells were located in the GI tract and activated by T-independent pathways. Hormonal regulation caused IgA to move to the mammary glands in the last trimester 43.
Antibodies have also been used to evaluate the safety and limitations of vaccines, specifically for potency and environmental contamination 26. In the development of Zika virus (ZIKV) vaccines they enabled the evaluation of subunit vaccines through a designed antigen for Zika specific mAbs. They noted that the antibodies specifically bind to the quaternary structures of the epitopes on the envelope (E) proteins. At physiological temperatures, the antigen favors the monomeric state which enables its evasion of antibody response. In response to this new information, they produced a dimerized E protein via disulfide bridge formation. This was tested against a variety of ZIKV mAbs using ELISA and only the dimerized antigen-stimulated the protective antibody response 44.
As we can see from these studies antibodies are a versatile tool in basic research and studies of antibodies themselves can explain human processes. Food allergies can be explained through studies of IgE. IgE undergoes class switching to other antibody classes. When class switched to IgG4 tolerance to foods was noted. When regulatory B cells are reduced IgE doesn’t switch to IgG4 affording no protection which is what happens in people with food allergies 45. Other studies of antibodies have concluded that the C region affects the affinity and specificity of the Ab as a whole and C regions of DNA-binding antibodies specifically impacted specificity and secondary structure formation 46. These studies can influence how antibodies are engineered to induce different functions and improve characteristics as well as showing mechanisms that can be useful for therapeutic development for diseases and disorders affected by antibodies.
Suppliers: Absolute Antibody, BioRad Antibodies, Creative BioMart
These testing techniques have enabled vast amounts of research using antibodies. In studies of neurobiology, autoantibodies have been noted to play a role in autoimmune neurological diseases particularly encephalitis. With the use of recombinant patient-derived mAbs, they determined how the autoantibodies work. They cross-link proteins on the synaptic cell surface such as neurotransmitters like N-methyl-D-aspartate (NMDA) receptors and cause internalization of these receptors affecting amino-terminal domain (ATD)-mediated gated control presenting as symptoms of epilepsy, psychosis, etc. IHC staining assisted this study by providing information about the localization of these autoantibodies and the Leucine-rich glioma-inactivated 1 (LGI1) protein was noted to affect protein-protein interactions. Autoantibody potential in basic neuroscience was discovered with further uses in structure-function relationships, discovery of protein-specific roles in vivo from induced recombinant antibody (rAb) expression, epitope mapping, and analysis of protein distribution as well as studies of its own role in the body 40.
Antibodies are notoriously beneficial in proteomic studies. Their ability to bind proteins with specific characteristics provides more insight into certain diseases such as autophagy. Autophagy is caused by phosphorylation of the Autophagy Related 16 Like 1 (ATG16L1) protein in response to cellular and metabolic stresses. Through a collaboration with Abcam, the researchers designed a mAb that focuses on ATG16L1 phosphorylation on serine 278 of newly formed autophagosomes in mammalian cells. It’s important that the autophagosomes are new as dysfunctional ones can skew experimental results. Their studies with this antibody using ATG16L1 knockout and wildtype versions suggest that the rate of autophagy is controlled by another regulatory event in an uncharacterized region. Along with starvation which increases the number of new autophagosomes formed, and other autophagy inducing stresses. The success of this antibody has seen it become commercially available to all researchers and is compatible with IHC, IF, and western blotting 41.
Antibodies are known for the immunity they afford us, much of this is passed on from mother to child via colostrum and breast milk. Different factors can have separate effects on the concentrations of Igs in colostrum. One study found that malnourishment results in only a third of normal IgG concentrations and identified reductions in IgA and C4 of the complement pathway of the immune response 42. Another such study looked at the effects of immunization and infection during pregnancy on IgA colostrum concentrations. Through the use of ELISA and western blot they measured the IgA subclasses; ELISA quantified concentrations of IgA in relation to the vaccine antigens, and western blot evaluated the ability of colostrum to recognize antigens of E.coli. IgA2 was more reactive against lipopolysaccharide (LPS) and increased in women with multiple gastrointestinal (GI) infections, IgA1 increased in response to multiple doses of tetanus vaccine. The ELISA analysis also identified an increase of IgA2 in relation to the Pneumococcal polysaccharides (PPS) vaccine. Previous studies suggested that IgA-producing cells were located in the GI tract and activated by T-independent pathways. Hormonal regulation caused IgA to move to the mammary glands in the last trimester 43.
Antibodies have also been used to evaluate the safety and limitations of vaccines, specifically for potency and environmental contamination 26. In the development of Zika virus (ZIKV) vaccines they enabled the evaluation of subunit vaccines through a designed antigen for Zika specific mAbs. They noted that the antibodies specifically bind to the quaternary structures of the epitopes on the envelope (E) proteins. At physiological temperatures, the antigen favors the monomeric state which enables its evasion of antibody response. In response to this new information, they produced a dimerized E protein via disulfide bridge formation. This was tested against a variety of ZIKV mAbs using ELISA and only the dimerized antigen-stimulated the protective antibody response 44.
As we can see from these studies antibodies are a versatile tool in basic research and studies of antibodies themselves can explain human processes. Food allergies can be explained through studies of IgE. IgE undergoes class switching to other antibody classes. When class switched to IgG4 tolerance to foods was noted. When regulatory B cells are reduced IgE doesn’t switch to IgG4 affording no protection which is what happens in people with food allergies 45. Other studies of antibodies have concluded that the C region affects the affinity and specificity of the Ab as a whole and C regions of DNA-binding antibodies specifically impacted specificity and secondary structure formation 46. These studies can influence how antibodies are engineered to induce different functions and improve characteristics as well as showing mechanisms that can be useful for therapeutic development for diseases and disorders affected by antibodies.
Suppliers: Absolute Antibody, BioRad Antibodies, Creative BioMart
3.2 Therapeutics
The homogeneity of mAbs, in contrast to the heterogeneity of pAbs, has enabled their use as therapeutics. The different forms of intrinsic activity caused by pAbs, including both neutralizing and non-neutralizing antibodies, have ruled them out for this application 47. The first US Food and Drug Administration (FDA) approval of a mAb for therapeutic use was in 1986. Muromonab-CD3 is a transplant rejection (immunosuppressant) therapy and was fully murine 48. In April of 2021, the FDA hit a landmark, approving their 100th mAb therapeutic 49.
4 Overcoming Immunogenicity
The immunogenicity of mouse-derived antibodies saw the transformation to chimeric, humanized, and fully human antibodies. The immunogenicity reveals itself in the form of a rapid human anti-mouse antibody (HAMA) response that can lead to allergic reactions.
4.1 Chimerization
Chimerization is performed by genetic engineering with a murine hybridoma. The mouse C domains are essentially swapped out for the human counterpart and the mouse V regions are retained for specificity. The hybridoma secretes the chimeric antibodies which are then purified for use 23. The first approved therapeutic chimeric antibody, abciximab, was produced in 1994 50. This mAb was for the treatment of cardiovascular disease and the success was quickly translated into the first anticancer mAb (rituximab), also chimeric, in 1997 51.
4.2 Humanization
Humanization can be subdivided into rational and empirical methods of production in murine hybridomas 52. The most commonly used method is rational CDR grafting. This involves defining the region to be grafted from the mouse antibody, deciding the sequence used to produce the human frame, and selecting the additional back mutation residues for antibody affinity. The CDR from the mouse H chain is grafted onto the human framework region, this then has the back mutations induced to allow for integration of the CDR into the framework, preventing affinity loss 52. The first humanized mAb, daclizumab, was approved for use in 1997, also an immunosuppressant of transplant rejection 53.
4.3 Fully Human
Fully human antibodies are generated via four methods: transgenic mice, the single B cell, transgenic plants, or by display techniques (including phage, yeast, and the ribosome).
The transgenic method involves the immunization of mice via injection of the specific antigen and an immunostimulant (adjuvant). The spleen is dissected and the isolated splenocytes are fused to myeloma cells to create the hybridoma. After 2 weeks, the hybridoma supernatants are harvested and screened for cloning with ELISA. RNA is extracted from the hybridoma. The L and H chain V region genes are amplified and assembled with a joining region. This is then put into the expression vector, transfected into the mammalian cells where the antibodies are produced and secreted 54. The first transgenically produced fully human mAb was approved for use in 2006. Panitumumab was designed for an anti-epidermal growth factor receptor (EGFR) function as an alternative to chemotherapy 55.
The aforementioned production techniques all require the use of animals which as previously mentioned is ethically concerning. They are associated with higher costs to reduce their immunogenicity and have a reduced affinity compared to natural human antibodies 56. These methods are predicted to be replaced with the following animal-free methods shortly, driven by changes in regulations and scientific backing of non-animal derived methods to produce human antibodies with better characteristics.
Of the display techniques, phage display is one of the most popular methods of recombinant antibody production. It was first developed at the University of Cambridge in 1990 57 and in 2018 the Nobel prize for chemistry was awarded for the development of phage display of both proteins and antibodies 58.
All display techniques are based on the use of antibody gene libraries that are packed into either phage, yeast, or ribosomes and express antibody fragments on their protein surface for binding to target antigens. For the M13 phage, this surface protein is the pIII minor coat protein, also known as G3P, which the antibody fragments form a complex with to be displayed 59. This phage is a part of the Ff group of phage that only infect E.coli. Other phages can be used also such as lambda and T4 phages 60. Antibody domains are displayed on their protein surfaces as Fab, single-chain variable fragment (scFv), and nanobody structures (single-domain antibodies composed of a heavy chain and a variable domain) 61.
Phagemid vectors can be used as an alternative to phages. They are classed as ‘minimal’ phages as they contain the basic elements of the normal phage but require a helper phage that contains the full M13 genome to allow them to show the antibody-G3P complexes to become functional. As they require additional steps and have alower diversity of antibody binders (when compared to phage), they have become more popular in library development than as a library vector 60.
With display libraries there are 4 to choose from; naïve (natural antibodies from human donors B cells, no immunization needed), semi-synthetic (natural antibodies with mutated CDRs), synthetic (scaffold sequences of CDRs with induced diversity), or the immune library (clones of antibody genes from immunized animals and humans) 59. They typically consist of 106 – 1011 antibodies 62. For the selection of therapeutic antibodies, the naïve phage library is the library of choice as the antibodies have been taken from healthy donor B cells therefore will already have tolerance and stability within the human body 63.
The transgenic method involves the immunization of mice via injection of the specific antigen and an immunostimulant (adjuvant). The spleen is dissected and the isolated splenocytes are fused to myeloma cells to create the hybridoma. After 2 weeks, the hybridoma supernatants are harvested and screened for cloning with ELISA. RNA is extracted from the hybridoma. The L and H chain V region genes are amplified and assembled with a joining region. This is then put into the expression vector, transfected into the mammalian cells where the antibodies are produced and secreted 54. The first transgenically produced fully human mAb was approved for use in 2006. Panitumumab was designed for an anti-epidermal growth factor receptor (EGFR) function as an alternative to chemotherapy 55.
The aforementioned production techniques all require the use of animals which as previously mentioned is ethically concerning. They are associated with higher costs to reduce their immunogenicity and have a reduced affinity compared to natural human antibodies 56. These methods are predicted to be replaced with the following animal-free methods shortly, driven by changes in regulations and scientific backing of non-animal derived methods to produce human antibodies with better characteristics.
Of the display techniques, phage display is one of the most popular methods of recombinant antibody production. It was first developed at the University of Cambridge in 1990 57 and in 2018 the Nobel prize for chemistry was awarded for the development of phage display of both proteins and antibodies 58.
All display techniques are based on the use of antibody gene libraries that are packed into either phage, yeast, or ribosomes and express antibody fragments on their protein surface for binding to target antigens. For the M13 phage, this surface protein is the pIII minor coat protein, also known as G3P, which the antibody fragments form a complex with to be displayed 59. This phage is a part of the Ff group of phage that only infect E.coli. Other phages can be used also such as lambda and T4 phages 60. Antibody domains are displayed on their protein surfaces as Fab, single-chain variable fragment (scFv), and nanobody structures (single-domain antibodies composed of a heavy chain and a variable domain) 61.
Phagemid vectors can be used as an alternative to phages. They are classed as ‘minimal’ phages as they contain the basic elements of the normal phage but require a helper phage that contains the full M13 genome to allow them to show the antibody-G3P complexes to become functional. As they require additional steps and have alower diversity of antibody binders (when compared to phage), they have become more popular in library development than as a library vector 60.
With display libraries there are 4 to choose from; naïve (natural antibodies from human donors B cells, no immunization needed), semi-synthetic (natural antibodies with mutated CDRs), synthetic (scaffold sequences of CDRs with induced diversity), or the immune library (clones of antibody genes from immunized animals and humans) 59. They typically consist of 106 – 1011 antibodies 62. For the selection of therapeutic antibodies, the naïve phage library is the library of choice as the antibodies have been taken from healthy donor B cells therefore will already have tolerance and stability within the human body 63.
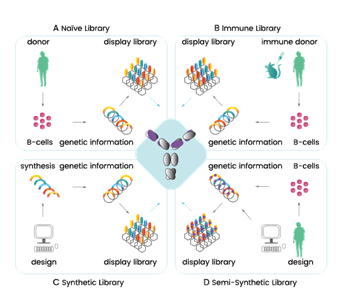
Figure 4.3.1: Illustration of the 4 types of display libraries, showing broadly how they differ in production 64.
To select antibodies specific to an antigen, a process called affinity selection is implemented. This involves the incubation of the phage library with the immobilized antigen target (a form of ELISA assay), the unbound antibodies are washed off to produce a sub-library. The phage is eluted from the immobilized antigen by altering the pH levels with buffering solution or by enzymatic digestion using trypsin or other acids/bases 59, 60. This elution step does more than just detach the antibody-antigen from the well, it also causes the bald phage, phage that hasn’t got the G3P-antibody complex on their surface, to become non-infectious reducing the background noise. The final step is to sequence and transfect the antibody genes into high yield expression vectors such as E.coli for large scale production in a host cell line 28, 65, a helper phage can sometimes be added to assist with amplification of the infective phage in the E.coli expression vector 60. These steps are repeated 1-5 times, starting with the products of the amplification step each time, to produce a sub-library with only the most targeted antibodies in it 28. After the antibodies of interest have been chosen they undergo an optimization step in which they are engineered, by light chain shuffling or other methods, to improve affinity, yield, and stability 63. Additionally, polyclonal phage ELISA assays of the sub-libraries can confirm the selection of high-affinity antibodies, and titration of phage output can be used to monitor progress in the procedure 60.The first fully human therapeutic produced in this way, without the use of animals, was approved by the FDA in 2002. It is called adalimumab and treats rheumatoid arthritis by blocking tumor necrosis factor (TNF)𝛼 66.
Additional factors can be included with the antigen to produce enriched sub-libraries with specific antigen targets. For example, the addition of a cofactor that induces a certain conformation can produce a sub-library targeted to that conformation of the antigen. Or the addition of a soluble competitor can assist in ruling out any cross-reactivity and can locate antibodies for site-specific phosphorylation 67, 63.
Additional factors can be included with the antigen to produce enriched sub-libraries with specific antigen targets. For example, the addition of a cofactor that induces a certain conformation can produce a sub-library targeted to that conformation of the antigen. Or the addition of a soluble competitor can assist in ruling out any cross-reactivity and can locate antibodies for site-specific phosphorylation 67, 63.
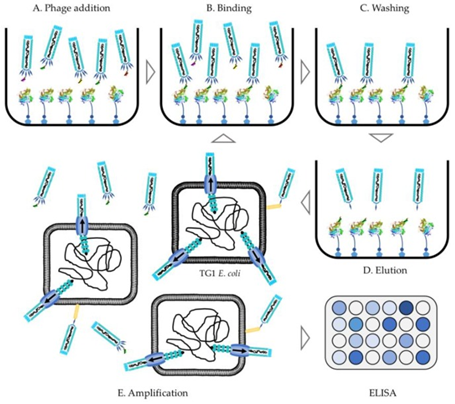
Figure 4.3.2: Flow diagram of the phage display protocol from library addition to the antigen of choice to ELISA conformation of antibody binding selection 60.
As of August 2020, there have been 14 phage display-produced mAbs approved for therapeutic use. More are available for research use with many labs choosing to create their own 68. Toxinology and antivenom research specifically use M13 phage display techniques for their studies 60.
Suppliers: Creative BioLabs, HybriBody, ProteoGenix, BioMolecular Discovery Services
Ribosomal display, first developed in 1994 69, differs in the library vector used. Instead of phage, the ribosome is the vector of choice. One of the four libraries mentioned above is used, their DNA is transcribed and then mRNA is translated by the ribosome, in vitro, to produce the antibody protein. In most cases, a complex of protein-ribosome-mRNA (PRM) is formed due to the absence of a terminal stop codon caused by a deleterious mutation that prevents the ribosome from detaching once translation is finished. These make up the ribosome antibody libraries that undergo selection in the same fashion as phage display, with an immobilized antigen. This complex allows for simultaneous selection of both the antibody protein and the mRNA code used to make it, ultimately reducing the procedure time. Then they are subjected to reverse transcription-polymerase chain reaction (RT-PCR) to recover the antibody encoding DNA. Once recovered the DNA is then amplified by PCR to allow for cloning, genetic manipulation, or the amplified DNA transcripts can undergo panning a further 3-5 times to generate a lead antibody for therapeutic evaluation 70, 71.
Additionally, the antibody clones can be tested by monoclonal ELISA, and affinity and stability can be increased by further maturation procedures. Ribosomal display has been particularly successful with affinity maturation protocols enabling correction of errors caused through DNA shuffling and RT-PCR 71.
Suppliers: Creative BioLabs, HybriBody, ProteoGenix, BioMolecular Discovery Services
Ribosomal display, first developed in 1994 69, differs in the library vector used. Instead of phage, the ribosome is the vector of choice. One of the four libraries mentioned above is used, their DNA is transcribed and then mRNA is translated by the ribosome, in vitro, to produce the antibody protein. In most cases, a complex of protein-ribosome-mRNA (PRM) is formed due to the absence of a terminal stop codon caused by a deleterious mutation that prevents the ribosome from detaching once translation is finished. These make up the ribosome antibody libraries that undergo selection in the same fashion as phage display, with an immobilized antigen. This complex allows for simultaneous selection of both the antibody protein and the mRNA code used to make it, ultimately reducing the procedure time. Then they are subjected to reverse transcription-polymerase chain reaction (RT-PCR) to recover the antibody encoding DNA. Once recovered the DNA is then amplified by PCR to allow for cloning, genetic manipulation, or the amplified DNA transcripts can undergo panning a further 3-5 times to generate a lead antibody for therapeutic evaluation 70, 71.
Additionally, the antibody clones can be tested by monoclonal ELISA, and affinity and stability can be increased by further maturation procedures. Ribosomal display has been particularly successful with affinity maturation protocols enabling correction of errors caused through DNA shuffling and RT-PCR 71.
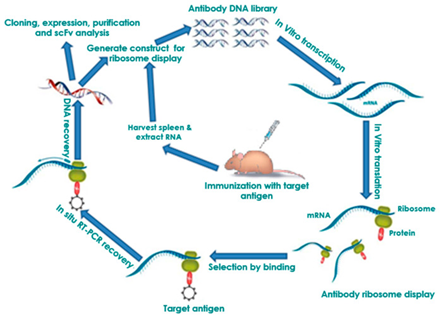
Figure 4.3.3: Flow diagram of the process of ribosomal display. The center steps involving the use of mice can be substituted with other antibody libraries such as naïve, semi-synthetic, synthetic, or immune (shown above) 71.
Ribosome display is part of the cell-free category of display techniques alongside mRNA display. The mRNA transcripts are translated in vitro. As a result of this, bigger ribosomal and mRNA display libraries have been created, in the 1012 to 1015 range 71, as they are not constrained by library size as in phage. This in turn gives more opportunity for individual preference, as there is a higher chance more than one antibody will bind allowing a choice of potencies 70.
It has become a crucial tool for the low-cost production of diagnostic antibodies and is very useful in research as, with the antibody sequence, the antibody protein can be created in different formats to suit different applications and created in different hosts for better compatibility. It has many advantages when compared with the cell-based methods, the most obvious is the lack of cell culture use which makes it more efficient. The cell-free methods of generation enable post-translational modification (PTM), allowing the introduction of genetic modifications to create antibodies with specific characteristics and to enable studies of the effect of PTMs. It also does not require compromisation of the library in the form of transformation which also negatively affects efficiency. By not compromising the library it allows for the use of larger libraries increasing the likelihood of finding a high-affinity binding antibody 71.
This display technique has been used in research to combat the ebola virus (EBV). It was used to develop a panel of single-chain antibodies against known EBV epitopes, allowing detection of different EBV species and Marburg virus associated with EBV, showing high diagnostic potential. It also produced antibodies against the Zika virus showing potential neutralizing and infection inhibiting abilities 71.
Suppliers: Creative BioLabs
Yeast display is part of the cell-based class, using the Saccharomyces yeast cells. It displays antibodies as scFv and Fab fragments with their H and L chains connected by a flexible polypeptide linker 62. It was first used for scFv fragments in 1997 72. In the yeast cells, the antibody fragments are situated on the a-agglutinin receptor bound to the C-terminus of the Aga2 protein, this protein is bound to the Aga1 protein by two disulfide bonds. Aga1 directly connects this complex to the yeast cell surface 73, 62. Recent studies have suggested that the connection of the antibody to the N-terminus of the Aga2 protein can overcome the limitations seen with nanobody display 73.
It has become a crucial tool for the low-cost production of diagnostic antibodies and is very useful in research as, with the antibody sequence, the antibody protein can be created in different formats to suit different applications and created in different hosts for better compatibility. It has many advantages when compared with the cell-based methods, the most obvious is the lack of cell culture use which makes it more efficient. The cell-free methods of generation enable post-translational modification (PTM), allowing the introduction of genetic modifications to create antibodies with specific characteristics and to enable studies of the effect of PTMs. It also does not require compromisation of the library in the form of transformation which also negatively affects efficiency. By not compromising the library it allows for the use of larger libraries increasing the likelihood of finding a high-affinity binding antibody 71.
This display technique has been used in research to combat the ebola virus (EBV). It was used to develop a panel of single-chain antibodies against known EBV epitopes, allowing detection of different EBV species and Marburg virus associated with EBV, showing high diagnostic potential. It also produced antibodies against the Zika virus showing potential neutralizing and infection inhibiting abilities 71.
Suppliers: Creative BioLabs
Yeast display is part of the cell-based class, using the Saccharomyces yeast cells. It displays antibodies as scFv and Fab fragments with their H and L chains connected by a flexible polypeptide linker 62. It was first used for scFv fragments in 1997 72. In the yeast cells, the antibody fragments are situated on the a-agglutinin receptor bound to the C-terminus of the Aga2 protein, this protein is bound to the Aga1 protein by two disulfide bonds. Aga1 directly connects this complex to the yeast cell surface 73, 62. Recent studies have suggested that the connection of the antibody to the N-terminus of the Aga2 protein can overcome the limitations seen with nanobody display 73.
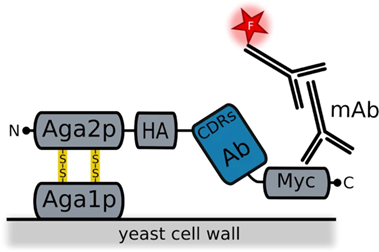
Figure 4.3.4: Image showing how the antibody is integrated into the yeast a-agglutinin surface receptor. The Aga1 protein is anchored to the cell surface. Aga2 is connected to Aga1 via two disulfide bonds. This Aga2 is also bound to a HA tag which the antibody fragment is attached to with a C-terminal Myc to finish off. Also shown in this diagram are the fluorescently labeled monoclonal antibodies used for detection 73.
For the antibody to be displayed on the a-agglutinin receptor it first has to be cloned into the yeast cells. The antibody gene is inserted into the yeast plasmid as an in-frame fusion with the Aga2 protein gene. This is then transformed into the yeast cell where the expression of the antibody gene is under the control of the GAL1 promoter, dependent on galactose for transcription and translation. When galactose is present the scFv antibodies are present in 104 – 105 copies on the yeast surface 72. Total size can be from 107 – 109 antibodies per library, this is smaller than the other libraries 62.
After cloning and expression of antibody fragments, the libraries are sorted by magnetic-activated cell sorting (MACS) to reduce the number of antibodies in the library to within 107 – 108, which is compatible with flow cytometry. The cells are then fluorescently labeled allowing monitoring of MACS before entering the flow cytometry process where it too can be used for monitoring expression 62. Areas with brighter fluorescence indicate high-affinity binding. Affinity can also be determined using equilibrium-based kinetic analysis 72. This process further enriches the MACS sorted cells, which are then checked for specificity and if needed can be engineered with random mutagenesis to increase affinity and stability 62. Mutagenesis in this case, causes transition and transversion mutations, and can also be called directed evolution. It allows the selection of antibody fragments with specific properties 62. To increase the diversity of the libraries DNA shuffling is implemented. For the engineering of individual scFvs, they must first be cloned into yeast surface display vectors 62.
Similar to ribosomal display, it also allows the choice of the highest affinity mAb because of the ability to measure binding affinity 74. The binding properties can be characterized without the need for subcloning, expression, and purification of the antibody fragments 72.
Yeast produces correctly folded antibodies due to their eukaryotic expression system. This is arguably better than E.coli for producing human antibodies as it enables PTMs and uses the eukaryotic endoplasmic reticulum and other machinery for protein folding 72, 62. This is advantageous as it reduces the production time ruling out the need for purification and testing of binding/efficacy as the antibody will be human-compatible 75.
Its compatibility with fluorescent-activated cell sorting (FACS) is also an advantage as it enables optical resolution distinguishing between cell types with fine resolution, can use fluorescence and light scattering for purification of heterogeneous cultures 72, and enables quantitative screening 62. Simultaneous stability and affinity engineering can also be achieved with yeast display libraries alongside two-color labeling fluorophores 62.
Yeast do have some downsides though, and the diversity of antibodies they display is lower than in phage display due to smaller library size 74, 76.
Suppliers: Creative BioLabs, AvantGen
The single B cell method involves PCR amplification of the human VH and VL encoding genes extracted from single human B cells. These are purified and cloned into an expression vector which is placed in mammalian cell culture lines to produce the antibody products 77. These products are screened using microarray and micro engraving technologies 78.One adapted version of the B cell method is called Ecobody technology and has applied human immortalized B cells to influenza virus vaccine research. It overcomes the limitations of the basic method through its use of a modified Fab format known as ‘Zipbody’ that has leucine zippers at the C-terminus of both L and H chains, overcoming the incorrect folding seen with normal Fab L and H chains. Its procedure consists of isolating B cells from human blood. For the influenza virus vaccine study this was specifically Epstein-Barr virus B cells. They are fluorescently tagged with an anti-human IgG to select for only IgG-producing cells. These B cells are then subjected to FACS and incubated with the antigens of interest. The cells bound to the beads are separated and are amplified by RT-PCR. Target proteins can be found in both the soluble and insoluble fractions, to extract them from the soluble fractions His-Tag purification is needed. These amplified genes are first put into vectors before finally being put into E.coli for expression. The final step is ELISA analysis of the expressed antibodies. The Ecobody technology has one striking advantage over conventional B cell methods; it can screen mAbs in only 2 days 79.
After cloning and expression of antibody fragments, the libraries are sorted by magnetic-activated cell sorting (MACS) to reduce the number of antibodies in the library to within 107 – 108, which is compatible with flow cytometry. The cells are then fluorescently labeled allowing monitoring of MACS before entering the flow cytometry process where it too can be used for monitoring expression 62. Areas with brighter fluorescence indicate high-affinity binding. Affinity can also be determined using equilibrium-based kinetic analysis 72. This process further enriches the MACS sorted cells, which are then checked for specificity and if needed can be engineered with random mutagenesis to increase affinity and stability 62. Mutagenesis in this case, causes transition and transversion mutations, and can also be called directed evolution. It allows the selection of antibody fragments with specific properties 62. To increase the diversity of the libraries DNA shuffling is implemented. For the engineering of individual scFvs, they must first be cloned into yeast surface display vectors 62.
Similar to ribosomal display, it also allows the choice of the highest affinity mAb because of the ability to measure binding affinity 74. The binding properties can be characterized without the need for subcloning, expression, and purification of the antibody fragments 72.
Yeast produces correctly folded antibodies due to their eukaryotic expression system. This is arguably better than E.coli for producing human antibodies as it enables PTMs and uses the eukaryotic endoplasmic reticulum and other machinery for protein folding 72, 62. This is advantageous as it reduces the production time ruling out the need for purification and testing of binding/efficacy as the antibody will be human-compatible 75.
Its compatibility with fluorescent-activated cell sorting (FACS) is also an advantage as it enables optical resolution distinguishing between cell types with fine resolution, can use fluorescence and light scattering for purification of heterogeneous cultures 72, and enables quantitative screening 62. Simultaneous stability and affinity engineering can also be achieved with yeast display libraries alongside two-color labeling fluorophores 62.
Yeast do have some downsides though, and the diversity of antibodies they display is lower than in phage display due to smaller library size 74, 76.
Suppliers: Creative BioLabs, AvantGen
The single B cell method involves PCR amplification of the human VH and VL encoding genes extracted from single human B cells. These are purified and cloned into an expression vector which is placed in mammalian cell culture lines to produce the antibody products 77. These products are screened using microarray and micro engraving technologies 78.One adapted version of the B cell method is called Ecobody technology and has applied human immortalized B cells to influenza virus vaccine research. It overcomes the limitations of the basic method through its use of a modified Fab format known as ‘Zipbody’ that has leucine zippers at the C-terminus of both L and H chains, overcoming the incorrect folding seen with normal Fab L and H chains. Its procedure consists of isolating B cells from human blood. For the influenza virus vaccine study this was specifically Epstein-Barr virus B cells. They are fluorescently tagged with an anti-human IgG to select for only IgG-producing cells. These B cells are then subjected to FACS and incubated with the antigens of interest. The cells bound to the beads are separated and are amplified by RT-PCR. Target proteins can be found in both the soluble and insoluble fractions, to extract them from the soluble fractions His-Tag purification is needed. These amplified genes are first put into vectors before finally being put into E.coli for expression. The final step is ELISA analysis of the expressed antibodies. The Ecobody technology has one striking advantage over conventional B cell methods; it can screen mAbs in only 2 days 79.
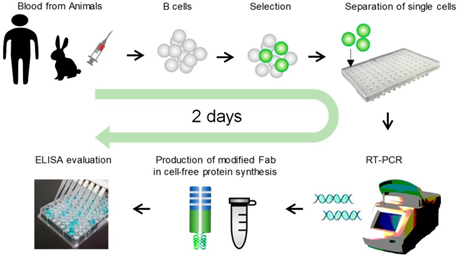
Figure 4.3.5: Image showing the use of Ecobody technology to screen monoclonal antibodies from human B cells 79.
The B cells can be isolated from the blood or bone marrow, by fluorescence-activated cell sorting or other techniques, which allows the identification of B cells at different stages of development. In general, donors have already recovered from infection caused by a specific antigen that produces the antibody of interest. This means these isolated B cells do not need artificial immunization as the antibody genes are already present 77, 80.
This technique, as with all techniques, has advantages and disadvantages. They don’t use animals for their production, but donors can be scarce due to the requirement for specific disease recovery and the antibodies are ineffective against certain molecules 77. However, in the case of this technique, the benefits outweigh the limitations. These antibodies have seen increased qualities such as; safety, efficacy, and immunotolerance 80.
Suppliers: GenScript ProBio, SinoBiological, Genovac
The transgenic plant method involves the transformation of human antibody genes into plants creating transgenic plants that will produce either full length antibodies or antibody fragments. Plants were first considered for antibody production in the 90s 81.There are a few different protocols for production. The conventional system sees the transformation of antibody-encoding genes into transient expression vectors which were then subsequently put into the plant embryo to grow. Once grown the antibodies can be extracted and purified 82.
The plant cell culture method sees plant cells in the differentiated state grown alongside the protein (obtained from biomass or culture liquid) in a bioreactor. This method of production is faster, more economical and introduces less contaminants 83.
Production in the tobacco plant occurs through sexual crossing in which two tobacco plants, one with the kappa chain transformed into it and the other with the gamma chain, cross to produce antibodies that contain both chains 84. Tobacco is the plant of choice for antibody production as it has the best yield per hectare as it can be cropped many times in one year. For example, 360 million doses of the anthrax antibody can be produced from one acre of tobacco 85. It is not a food or feed so can be grown in polytunnels where the risk of cross-contamination is minimal and it will not result in a food shortage 82.
Although tobacco is the plant of choice, researchers recommend the production of antibodies in seeds, as plants can be poor long term storage hosts as their proteases cause degradation. Seeds on the other hand have very low concentrations of proteases which have minimal effects on the bioactivity of the antibody 86. Plants are engineered to express the antibodies in their seeds with the use of seed specific regulatory sequences 87.
Plantibodies have distinct advantages over the use of cell lines for the production of antibodies. They are plentiful and can mature in only one season meaning production is quick and costs less than other methods 88. Plants are less likely to introduce pathogens to the antigens than mammalian cells meaning that screening for pathogens is not necessary as plants and humans share similar endomembrane systems and secretory pathways 89. Large scale production is already in place for many plants such as corn, for example, meaning that up-scaling of plantibodies can occur quickly 90. Long term storage with little effects on bioactivity can happen when plantibodies are stored in seeds 88, 85. Plants are also capable of glycosylating proteins which provides resistance to thermal degradation, increases the immunogenic power, and protects against proteolytic degradation 87.
This technique, as with all techniques, has advantages and disadvantages. They don’t use animals for their production, but donors can be scarce due to the requirement for specific disease recovery and the antibodies are ineffective against certain molecules 77. However, in the case of this technique, the benefits outweigh the limitations. These antibodies have seen increased qualities such as; safety, efficacy, and immunotolerance 80.
Suppliers: GenScript ProBio, SinoBiological, Genovac
The transgenic plant method involves the transformation of human antibody genes into plants creating transgenic plants that will produce either full length antibodies or antibody fragments. Plants were first considered for antibody production in the 90s 81.There are a few different protocols for production. The conventional system sees the transformation of antibody-encoding genes into transient expression vectors which were then subsequently put into the plant embryo to grow. Once grown the antibodies can be extracted and purified 82.
The plant cell culture method sees plant cells in the differentiated state grown alongside the protein (obtained from biomass or culture liquid) in a bioreactor. This method of production is faster, more economical and introduces less contaminants 83.
Production in the tobacco plant occurs through sexual crossing in which two tobacco plants, one with the kappa chain transformed into it and the other with the gamma chain, cross to produce antibodies that contain both chains 84. Tobacco is the plant of choice for antibody production as it has the best yield per hectare as it can be cropped many times in one year. For example, 360 million doses of the anthrax antibody can be produced from one acre of tobacco 85. It is not a food or feed so can be grown in polytunnels where the risk of cross-contamination is minimal and it will not result in a food shortage 82.
Although tobacco is the plant of choice, researchers recommend the production of antibodies in seeds, as plants can be poor long term storage hosts as their proteases cause degradation. Seeds on the other hand have very low concentrations of proteases which have minimal effects on the bioactivity of the antibody 86. Plants are engineered to express the antibodies in their seeds with the use of seed specific regulatory sequences 87.
Plantibodies have distinct advantages over the use of cell lines for the production of antibodies. They are plentiful and can mature in only one season meaning production is quick and costs less than other methods 88. Plants are less likely to introduce pathogens to the antigens than mammalian cells meaning that screening for pathogens is not necessary as plants and humans share similar endomembrane systems and secretory pathways 89. Large scale production is already in place for many plants such as corn, for example, meaning that up-scaling of plantibodies can occur quickly 90. Long term storage with little effects on bioactivity can happen when plantibodies are stored in seeds 88, 85. Plants are also capable of glycosylating proteins which provides resistance to thermal degradation, increases the immunogenic power, and protects against proteolytic degradation 87.
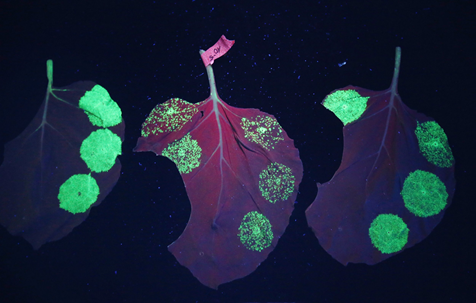
Figure 4.3.6: Nicotiana benthamiana part of the Nicotine plant family with fluorescent spots of green fluorescent protein (GFP) indicating the presence of plantibodies 91, 92.
Newer plant transformation technologies have since come about to strengthen protein production in plants. This includes plastid engineering where the gene is put into a plastid rather than the genome as plastids are more abundant which enables increased expression 87. Agroinfiltration is a method of inducing transient expression with the use of agrobacterium as the vector for delivery with vacuum infiltration 93. Magnifection, is also a transient expression system which uses agrobacterium to deliver viral vectors for high level expression and deconstructed viral vectors can be used to prevent movement from cell to cell 87.
Suppliers: Planet Biotechnology, Leaf Expression Systems, Icon Genetics
There are other expression vectors being tested for recombinant antibodies including insects such as Drosophila melanogaster, gram positive bacteria such as Bacillus subtilis, fungi such as Aspergillus, and protozoa that can perform both N- and O-glycosylation 94.
Suppliers: Planet Biotechnology, Leaf Expression Systems, Icon Genetics
There are other expression vectors being tested for recombinant antibodies including insects such as Drosophila melanogaster, gram positive bacteria such as Bacillus subtilis, fungi such as Aspergillus, and protozoa that can perform both N- and O-glycosylation 94.
5 Advantages of Animal-Free Antibodies (AFAs)
Animal-free antibodies have several advantages over traditional animal-produced antibodies. They play a key role in the third R of the 3R system; replacement. By replacing animals as the expression vectors, with phage, yeast, or single B cells, for antibody discovery, and mammalian cell lines for production, not only reduces the number of animals used in science but also increases the specificity and reduces the immunogenicity of antibodies.Hybridomas can be unreliable. They are subject to contamination, genetic drift and their instability can lead to loss of antibody expression or additional expression of L chains 27. Hybridomas are dependent on animal-derived growth factors and the switch to serum-free growth was not compatible. To reduce and replace animals in research we need to steer clear of hybridoma reliant methods 95. AFAs on the other hand have little batch-to-batch variation and are more reproducible making them an excellent alternative 31.Inclusion of competitor molecules in the production of AFAs has enabled direct selection of specific targets 26. The affinity of antibodies produced in this way is much higher than immunization and allows control over both selection and screening. Otherwise conserved antibody targets are accessible through the use of in vitro methods, this was previously not feasible due to immunological tolerance. In vitro methods do not need additional steps to overcome this tolerance whereas immunization requires extra steps to overcome their immunogenicity lengthening an already long process.
As the antibody sequence is known and can be cloned it has made the process more reproducible. It has also allowed the manipulation and editing of genes to produce new antibodies and to select for the production of only certain fragments. Antibodies can also be selected for certain characteristics through the inclusion of competitor molecules or agents to induce certain conformations. Recombinant antibodies are valuable tools in the lab as they can be converted to species compatible forms by changing the C domain to the native species’ 28.
Hybridomas were quickly established as a valid method of antibody production and with high uptake, uptake of phage display on the other hand was slower. This lag was mainly due to patents, many of these patents have started to expire in recent years seeing an increase in the use of phage technologies which they expect to rival the uptake levels seen for hybridomas 76. AFAs have been well established in the therapeutic sector but uptake in other sectors hasn’t matched up 96.
Phage display has the advantage that it requires less antigen to locate a matching antibody than immunization, while also using controlled selection conditions that allow for smaller fragment production along with allowing the swapping of fragments 26.In vitro technologies can identify differences in protein sequences and structural changes in the small molecules they target. Phage display is uniquely good at identifying tyrosine sulfation, a common PTM that is responsible, in part, for protein-protein interactions and other functions in the body. It is generally hard to target but phage display allows the selection of antibodies specific enough to recognize them 76, 97.
One big advantage of AFAs over animal-derived is the ability to increase affinity through the use of more than one library, creating smaller and smaller clonotypes at the end of each new library, leaving the highest affinity antibodies to choose from in the last clonotype.Through in vitro methods ELISA positive antibodies can be generated in as little as 2 weeks with an average production time taking 8 weeks. The average time for hybridoma-generated antibodies is 4 months 28. In addition to this, large-scale production of rAbs is much cheaper making them more accessible to researchers 98.
These are constantly evolving technologies and over the years previous limitations have been overcome such as, the production of unglycosylated or incorrect glycosylation of antibodies from bacterial or yeast cultures which were swapped out for human cell lines producing fully human antibodies in vitro.
These advantages make a very convincing argument, and the EU parliament agreed. In 2010 a law (2010/63/EU) 99 was passed that prohibits the production of antibodies through animal methods where alternative methods with equal or better quality exist. This is just one part of the EU’s commitment to the 3R process of animal use, another part includes the ban on cosmetic testing on animals. This quickly spread to other countries due to the inclusion of imported cosmetics under this law. The 2010/63/EU law also discourages the use of imported animal-derived antibodies 96, which has seen talks overseas in the Organization for Economic Co-operation and Development (OECD) and the US National Institutes of Health (NIH) as to what they can do to prevent a trade stop 100.
This law has been a long time coming after the EURL ECVAM safety advisory (ESAC) advised in 1998 that the in vivo production of mAbs in animals, specifically through the ascites method was no longer needed as its alternative, phage display produced antibodies, provided as good as, if not better quality reagents than the ascites method. This had the added advantage of also reducing cruelty associated with traditional production methods 101. A minimum of 2 rabbits or guinea pigs and between 5-10 rats or mice are used in polyclonal production methods and approximately 5 mice are immunized for every one hybridoma produced. These all make up a sizable chunk of the 2.7 million animals used for science and medicine in the EU. Under this law, production or testing via animal methods requires a very valid and robust justification that has to be supplied in the application and approved by the authorities before proceeding 100.
There is no scientific justification for the use of animals to produce antibodies. Phage displayed antibodies don’t just look like in vivo antibodies, they also have the same functional biological mechanisms as the in vivo adaptive immune system. They are identical to in vivo antibodies in both structure and function so meet the requirements to support the 2010/63/EU directive 100.
We have found ourselves in a reproducibility crisis, in one such study a mere 6 of 53 studies were reproducible and in a separate study they found that less than 3000 of some 6000 commercial antibodies only bound their specified target, meaning more than half of those antibodies bound additional off-target epitopes or didn’t bind their specified target at all. Many commercially produced antibodies are not validated and as a result many antibodies can have low specificity for their target antigen. Most large pharmaceutical companies have departments dedicated to validation but the same cannot be said for research antibodies. In most research papers, the source of antibodies is not stated, and when they do, purchasing the same antibodies does not guarantee the same batch and target specificity. As mentioned before batch variation is a big reproducibility issue with animal derived antibodies, each new immunization will create a new mix of antibodies so it is impossible to get the same batch twice unless from the same animal or by recombinant animal-free methods 102.
In this report they recommend two ways to make antibodies more reproducible. The first is by defining antibodies by sequence and producing them recombinantly. Phasing out of hybridoma-derived antibodies is possible by sequencing the popular antibodies and making them recombinantly, preventing the future immunization of animals. The second recommendation was to use methods such as the display methods or single B-cell technology which use controlled protocols 102.
More advantages can be found on the People for Ethical Treatment of Animals (PETA) website here, including ethical and economic reasons for changing to AFAs.
As the antibody sequence is known and can be cloned it has made the process more reproducible. It has also allowed the manipulation and editing of genes to produce new antibodies and to select for the production of only certain fragments. Antibodies can also be selected for certain characteristics through the inclusion of competitor molecules or agents to induce certain conformations. Recombinant antibodies are valuable tools in the lab as they can be converted to species compatible forms by changing the C domain to the native species’ 28.
Hybridomas were quickly established as a valid method of antibody production and with high uptake, uptake of phage display on the other hand was slower. This lag was mainly due to patents, many of these patents have started to expire in recent years seeing an increase in the use of phage technologies which they expect to rival the uptake levels seen for hybridomas 76. AFAs have been well established in the therapeutic sector but uptake in other sectors hasn’t matched up 96.
Phage display has the advantage that it requires less antigen to locate a matching antibody than immunization, while also using controlled selection conditions that allow for smaller fragment production along with allowing the swapping of fragments 26.In vitro technologies can identify differences in protein sequences and structural changes in the small molecules they target. Phage display is uniquely good at identifying tyrosine sulfation, a common PTM that is responsible, in part, for protein-protein interactions and other functions in the body. It is generally hard to target but phage display allows the selection of antibodies specific enough to recognize them 76, 97.
One big advantage of AFAs over animal-derived is the ability to increase affinity through the use of more than one library, creating smaller and smaller clonotypes at the end of each new library, leaving the highest affinity antibodies to choose from in the last clonotype.Through in vitro methods ELISA positive antibodies can be generated in as little as 2 weeks with an average production time taking 8 weeks. The average time for hybridoma-generated antibodies is 4 months 28. In addition to this, large-scale production of rAbs is much cheaper making them more accessible to researchers 98.
These are constantly evolving technologies and over the years previous limitations have been overcome such as, the production of unglycosylated or incorrect glycosylation of antibodies from bacterial or yeast cultures which were swapped out for human cell lines producing fully human antibodies in vitro.
These advantages make a very convincing argument, and the EU parliament agreed. In 2010 a law (2010/63/EU) 99 was passed that prohibits the production of antibodies through animal methods where alternative methods with equal or better quality exist. This is just one part of the EU’s commitment to the 3R process of animal use, another part includes the ban on cosmetic testing on animals. This quickly spread to other countries due to the inclusion of imported cosmetics under this law. The 2010/63/EU law also discourages the use of imported animal-derived antibodies 96, which has seen talks overseas in the Organization for Economic Co-operation and Development (OECD) and the US National Institutes of Health (NIH) as to what they can do to prevent a trade stop 100.
This law has been a long time coming after the EURL ECVAM safety advisory (ESAC) advised in 1998 that the in vivo production of mAbs in animals, specifically through the ascites method was no longer needed as its alternative, phage display produced antibodies, provided as good as, if not better quality reagents than the ascites method. This had the added advantage of also reducing cruelty associated with traditional production methods 101. A minimum of 2 rabbits or guinea pigs and between 5-10 rats or mice are used in polyclonal production methods and approximately 5 mice are immunized for every one hybridoma produced. These all make up a sizable chunk of the 2.7 million animals used for science and medicine in the EU. Under this law, production or testing via animal methods requires a very valid and robust justification that has to be supplied in the application and approved by the authorities before proceeding 100.
There is no scientific justification for the use of animals to produce antibodies. Phage displayed antibodies don’t just look like in vivo antibodies, they also have the same functional biological mechanisms as the in vivo adaptive immune system. They are identical to in vivo antibodies in both structure and function so meet the requirements to support the 2010/63/EU directive 100.
We have found ourselves in a reproducibility crisis, in one such study a mere 6 of 53 studies were reproducible and in a separate study they found that less than 3000 of some 6000 commercial antibodies only bound their specified target, meaning more than half of those antibodies bound additional off-target epitopes or didn’t bind their specified target at all. Many commercially produced antibodies are not validated and as a result many antibodies can have low specificity for their target antigen. Most large pharmaceutical companies have departments dedicated to validation but the same cannot be said for research antibodies. In most research papers, the source of antibodies is not stated, and when they do, purchasing the same antibodies does not guarantee the same batch and target specificity. As mentioned before batch variation is a big reproducibility issue with animal derived antibodies, each new immunization will create a new mix of antibodies so it is impossible to get the same batch twice unless from the same animal or by recombinant animal-free methods 102.
In this report they recommend two ways to make antibodies more reproducible. The first is by defining antibodies by sequence and producing them recombinantly. Phasing out of hybridoma-derived antibodies is possible by sequencing the popular antibodies and making them recombinantly, preventing the future immunization of animals. The second recommendation was to use methods such as the display methods or single B-cell technology which use controlled protocols 102.
More advantages can be found on the People for Ethical Treatment of Animals (PETA) website here, including ethical and economic reasons for changing to AFAs.
6 Next-Generation Antibody Generation
Antibody discovery and production is an ever-evolving process with a near-constant flow of new techniques and technologies to advance antibody production. In this section, we will cover some of these advances including the incorporation of artificial intelligence (AI) and the production of bispecific antibodies.
6.1 Artificial Intelligence
AI has been employed for both discovery and generation through the use of its various types. Machine learning comes up frequently due to the impressive increase in affinity associated with its use. Long short-term memory (LSTM) specifically has been used in this application, identifying higher affinity antibody sequences for antibody affinity maturation. It was trained with next-generation sequencing (NGS) binding sequences and applied to anti-hapten antibody screening (also known as anti-target metabolite antibodies) to make better affinity antibodies. Compared with the anti-hapten screening on its own, the antibodies produced had 100-fold better affinity through exploration of virtual sequences beyond what is found in phage libraries 103.
Panning enrichment is an application of the convolutional neural network (CNN)-based regression model. It was noted that sequences from machine learning had higher affinity than panning methods 104. Through the use of both CNN and LSTM, they have been able to train models to distinguish between non-binder and binders for a clustered regularly interspaced short palindromic repeat (CRISPR) induced homology-directed repair (HDR) mechanism. This method has a hypothesized use in the optimization and engineering of therapeutic antibodies 105.
Deep learning, on the other hand, learns the long-range sequence dependencies. These dependencies in antibodies present as two amino acids separated by a long intermediate sequence but in 3D space are very close together. This makes it a very good platform for the identification of antibodies with correct binding sites and properties, as binding is generally based on shape. It is challenged by the size of the sets used for training, these are smaller than the sets they aim to build 106.
These are not the only applications, combinations of phage/yeast display followed by high throughput screening (HTS) and machine learning can create focused and antigen-specific clonotypes giving a choice of antibody affinity for the target antigen 27. In general, machine learning in the early stages of antibody discovery can be very useful for screening of function and affinity 106. AI has the potential to combine both in vivo and in vitro methods of production solving both ethical and scientific issues through generation with their vast libraries of antibodies including data on their structure, sequence, and binding affinity 107. Advances can bypass antibody libraries and move discovery to in silico models.
Panning enrichment is an application of the convolutional neural network (CNN)-based regression model. It was noted that sequences from machine learning had higher affinity than panning methods 104. Through the use of both CNN and LSTM, they have been able to train models to distinguish between non-binder and binders for a clustered regularly interspaced short palindromic repeat (CRISPR) induced homology-directed repair (HDR) mechanism. This method has a hypothesized use in the optimization and engineering of therapeutic antibodies 105.
Deep learning, on the other hand, learns the long-range sequence dependencies. These dependencies in antibodies present as two amino acids separated by a long intermediate sequence but in 3D space are very close together. This makes it a very good platform for the identification of antibodies with correct binding sites and properties, as binding is generally based on shape. It is challenged by the size of the sets used for training, these are smaller than the sets they aim to build 106.
These are not the only applications, combinations of phage/yeast display followed by high throughput screening (HTS) and machine learning can create focused and antigen-specific clonotypes giving a choice of antibody affinity for the target antigen 27. In general, machine learning in the early stages of antibody discovery can be very useful for screening of function and affinity 106. AI has the potential to combine both in vivo and in vitro methods of production solving both ethical and scientific issues through generation with their vast libraries of antibodies including data on their structure, sequence, and binding affinity 107. Advances can bypass antibody libraries and move discovery to in silico models.
6.2 Bispecific Antibodies
Bispecific antibodies are antibodies that have different H chain and different L chain pairs. They can be symmetric or asymmetric in structure and are classified based on this and their composition 21. Within the asymmetric and symmetric structure categories, there are also the Fc (IgG-like) and non-Fc (non-IgG) containing bispecifics. These categories determine the properties and applications of the antibody, for example, non-Fc containing bispecifics won’t be able to contribute to Fc-mediated effector functions such as ADCC, complement-dependent cytotoxicity (CDC), and antibody-dependent cellular phagocytosis (ADCP) but are smaller in size so will have enhanced penetration powers and reduced nonspecific recruitment 21, 108. They consist of two fragments, usually scFvs, held together by a peptide linker, disulfide bond, or non-covalent inter-domain interactions 109. There are also more than 100 other different bispecific antibody formats 109.
They can target either two different antigens at a time or two different epitopes of the one antigen which makes for a more specific, tailored treatment compared to natural antibodies. This can come in the form of, for example, simultaneously binding tumor cell receptors while also recruiting cytotoxic immune cells 19.There is no single standard production method, rather a variety of them that can produce the 100s of different bispecific antibody formats. Generation can be through genetic, biochemical, or molecular means. Production of first-generation bispecifics started in the 60s 110 through chemical conjugation of mAbs or by fusion of hybridomas. Chemical conjugation uses catalytic antibodies to join the reactive bispecific peptides of two different antibodies together, either whole Igs or only Fab fragments 111. Hybrid hybridomas (also known as quadromas) are created through somatic hybridization of two hybridomas 111. They generate cell lines that make two different H and L chains in a single cell. It wasn’t until the 90s that bispecifics entered their first clinical trials 108. Bispecifics at that time had high immunogenicity due to production using mouse antibodies which also induced a HAMA response reducing efficacy and inducing severe adverse effects 108, 111.
Advances over the years especially in rAb generation have been transferred to bispecific manufacturing of second-generation antibodies with defined properties and structure. This allowed the use of only the binding fragments of the antibody in bispecific production 111. Bispecifics are useful as therapeutics, initially only involved in effector cell retargeting but now also involved in dual targeting and pre-targeting, half-life extension, and delivery through cellular barriers among others 21. Large-scale production of bispecific antibodies has been established with the use of different engineering technologies such as asymmetric re-engineering technology (ART)-Ig 112. To create bispecifics with specific targets, engineering must enable the inclusion, for example, of the cluster of differentiation (CD)64 protein for the recruitment of macrophage and monocytes or CD16 to target NK cells and macrophage 108.
Bispecific antibodies can increase binding, selectivity, and anti-tumor activity to name but a few 111. As of 2018, there were only two bispecific antibodies approved for the treatment of cancer. An Fc-containing one, catumaxomab (now withdrawn in the EU 113), and a non-Fc containing one, blinatumomab. These are second-generation antibodies with either a combination of an anti-CD3 domain or an anti-Fc binding domain with anti-tumor binding domains. These bispecifics recruit immune effector cells for immunotherapy 108. In 2021 another was approved for use by the FDA for non-small cell lung cancer (NSCLC) called amivantamab 114. Only one other bispecific antibody has been approved for use, which is emicizumab for the treatment of hemophilia-A in 2018 115, but 100s of others are in development 116.
Both bispecific antibodies and antibody-drug conjugates (ADC) were created to overcome the resistance that has developed for normal therapeutic antibodies. ADCs consist of an antibody to target the antigens present on cancer cells and a potent cytotoxic payload that is connected by a chemical linker. After antibody binding, the ADC is endocytosed into the cell where the antibody or linker is degraded releasing the drug. This method has allowed the directed delivery of the drug into the cancer cell itself while sparing the normal healthy tissue 108.
They can target either two different antigens at a time or two different epitopes of the one antigen which makes for a more specific, tailored treatment compared to natural antibodies. This can come in the form of, for example, simultaneously binding tumor cell receptors while also recruiting cytotoxic immune cells 19.There is no single standard production method, rather a variety of them that can produce the 100s of different bispecific antibody formats. Generation can be through genetic, biochemical, or molecular means. Production of first-generation bispecifics started in the 60s 110 through chemical conjugation of mAbs or by fusion of hybridomas. Chemical conjugation uses catalytic antibodies to join the reactive bispecific peptides of two different antibodies together, either whole Igs or only Fab fragments 111. Hybrid hybridomas (also known as quadromas) are created through somatic hybridization of two hybridomas 111. They generate cell lines that make two different H and L chains in a single cell. It wasn’t until the 90s that bispecifics entered their first clinical trials 108. Bispecifics at that time had high immunogenicity due to production using mouse antibodies which also induced a HAMA response reducing efficacy and inducing severe adverse effects 108, 111.
Advances over the years especially in rAb generation have been transferred to bispecific manufacturing of second-generation antibodies with defined properties and structure. This allowed the use of only the binding fragments of the antibody in bispecific production 111. Bispecifics are useful as therapeutics, initially only involved in effector cell retargeting but now also involved in dual targeting and pre-targeting, half-life extension, and delivery through cellular barriers among others 21. Large-scale production of bispecific antibodies has been established with the use of different engineering technologies such as asymmetric re-engineering technology (ART)-Ig 112. To create bispecifics with specific targets, engineering must enable the inclusion, for example, of the cluster of differentiation (CD)64 protein for the recruitment of macrophage and monocytes or CD16 to target NK cells and macrophage 108.
Bispecific antibodies can increase binding, selectivity, and anti-tumor activity to name but a few 111. As of 2018, there were only two bispecific antibodies approved for the treatment of cancer. An Fc-containing one, catumaxomab (now withdrawn in the EU 113), and a non-Fc containing one, blinatumomab. These are second-generation antibodies with either a combination of an anti-CD3 domain or an anti-Fc binding domain with anti-tumor binding domains. These bispecifics recruit immune effector cells for immunotherapy 108. In 2021 another was approved for use by the FDA for non-small cell lung cancer (NSCLC) called amivantamab 114. Only one other bispecific antibody has been approved for use, which is emicizumab for the treatment of hemophilia-A in 2018 115, but 100s of others are in development 116.
Both bispecific antibodies and antibody-drug conjugates (ADC) were created to overcome the resistance that has developed for normal therapeutic antibodies. ADCs consist of an antibody to target the antigens present on cancer cells and a potent cytotoxic payload that is connected by a chemical linker. After antibody binding, the ADC is endocytosed into the cell where the antibody or linker is degraded releasing the drug. This method has allowed the directed delivery of the drug into the cancer cell itself while sparing the normal healthy tissue 108.
7 Conclusion
Antibodies are hugely beneficial in science and medicine, from their participation in assays to their use as treatments for the likes of cancer, rheumatoid arthritis, hemophilia, etc. These applications have benefited from advances in recent years that have enabled quicker production and production with specific advantages such as increased affinity or reduced immunogenicity making more specific antibodies with less chance of adverse reactions. These advances have also created choices for researchers with many variations of antibodies available from chimeric to fully human, covering all aspects of research and development. They have overcome many limitations over the years from immunogenicity and resistance to poor therapeutic targeting and efficacy. The creation of increased valency antibodies has enabled targeting to multiple epitopes ensuring the correct antigen is bound. Animal-derived methods of production are outdated and very rarely needed, AFAs are the way forward, they are generated in a fraction of the time, have the same if not better efficacy, reduced immunogenicity, and are ethically produced. There is the potential and the means to completely replace animals in the production and testing of antibodies and antibody products, which in years to come is what will be seen thanks to the cooperation of participating countries in the 3R (Reduction, Replacement, and Refinement) project for animal use in science, and the advances in science enabling this move.
8 Providers
The following providers either provide exclusively non-animal produced antibodies or animal free antibodies in addition to animal-derived antibodies.
Information provided courtesy of companies’ websites. Accio Biobank Online is not responsible for any information about, or services provided by, external companies.
Abcalis – By using our sequence defined recombinant antibodies, you can obtain superior quality of results with animal-free reagents. It is our mission and aspiration every single day to achieve this goal, both for a sustainable future and medical safety of this and every future generation to come.
Abcam – They have a selection of animal-free antibodies and proteins generated in E.coli, yeast, synthetic, human embryonic kidney (HEK), or Chinese hamster ovary (CHO) cell expression systems ranging from fragments to full-length proteins. They no longer use the ascites method but instead use tissue culture and expression vectors. The antibodies they provide are for research purposes only but they can also be used commercially through a partnership with Abcam.
Absolute Antibody – Absolute Antibody provides Antibody Sequencing, Engineering & Recombinant Expression.
Adimab – Discovery of antibodies is via a yeast system with either synthetic or natural libraries incorporated. This system allows the selection of properties in real-time. They can also use phage display libraries and can optimize almost any property. They also offer the ability to create bispecifics based on IgG, scFv, and CD3.
AdipoGen Life Sciences – Through the use of human naïve phage display libraries they produce animal-free monoclonal antibodies. Their antibodies focus on functional activation/blocking of their targets and they also employ a selection strategy that allows the selection of antibodies based on custom functions such as targeting a specific conformational state antigen or identification of multi-protein complexes. They are available via AdipoGen’s custom services for research, pre-clinical and therapeutic applications.
Aptagen – Aptagen is a leading world-wide biotechnology company offering aptamer (synthetic antibody) products and services as research reagents, diagnostic and biomarker discovery tools, as well as for use in drug discovery and targeted delivery for therapeutics, and bioindustrial applications. We have a combined 25+ years of experience in creating aptamers that bind a wide variety of targets against small molecules, proteins, biomarkers and cells. Aptagen has performed this service for more than 50 companies, organizations, and institutions worldwide.
AvantGen – AvantGen has constructed 30 human antibody libraries (Germliner™) with multiple human antibody germline frameworks. These were selected based on their high stability, solubility and expression level. Diversity is focused in the CDR regions and mimics the diversity of the natural human antibody repertoire for that variable region family. The framework sequences are left as fully human germline sequences. AvantGen’s proprietary libraries are comprised of >100 billion members, are well expressed and displayed with AvantGen’s yeast display system allowing efficient human antibody discovery.
BioRad – These have produced thousands of completely animal-free antibodies. They mainly use phage display with a fully synthetic, naïve human combinatorial antibody library. Recently they have employed a new technique of antibody production called SpyTag that makes antibodies by protein ligation and site-directed conjugation within 8 weeks. With each antibody webpage, they have included a list of applications tested in-house and through information in peer-reviewed journals.
Cell Signalling Technology (CST) – They also dropped the ascites method in favor of cDNA and phage libraries expressed in vectors such as CHO cells, HEK cells, Pichia pastoris, etc. CST only provides recombinant antibodies for research and development use.
Geneva Antibody Facility – Providing recombinant antibodies to the academic community, not-for-profit. It is also known as the ABCD database, each antibody within this database is linked to the UniProt website for more information as well as its own ABCD detailed summary which includes all known research applications, this is constantly being updated as new information is published. It is also linked to a manufacturing company that can make these antibodies on demand. It consists of over 23,000 antibodies and 4000 antigens.
GenScript – GenScript offers one non-animal expression method using bacterial vectors and a variety of recombinant antibodies for research use. Their BacPower system uses E.coli as the expression vector and can produce proteins in an average of 4-6 weeks. They also offer another alternative to animal use in their insect expression system. This system infects baculovirus-infected cells with the recombinant plasmid to produce either virus or recombinant antibodies.
Human Antibody Core Facility – One of the few laboratories in the world that produces fully-human, full-length, antigen-specific antibodies for use in studying human immune responses. The Core has achieved breakthroughs in antibody technology and produced hundreds of high affinity protective antibodies to influenza, anthrax lethal toxin, and various S. pneumonia polysaccharides. The Core aims to support investigators by helping quantify the antibody secreting cell responses after vaccination and by generating human monoclonal antibodies to be characterized. Pathogen-specific human monoclonal antibodies are also available for licensing agreements and other forms of commercial development.
HYBRiBODY – They produce antibodies through phage display offering selection of recombinant antibodies and nanobodies, that are ELISA validated, for basic research and diagnostics. They also offer additional services to test the validated antibodies including cell imaging and can create ‘minibodies’ with the species-specific Fc region you require. They can employ Yeast two Hybrid technology after successive rounds of phage display to screen for intrabodies, which are antibodies that work inside living cells. Intrabodies are very useful for live cell imaging and in vivo applications.
ProMab – They provide a library of human Fab fragments only, isolated from 120 donors. These antibodies are present in their general recombinant and natural catalogs but more specific antibodies can be obtained from their custom services too. They pan the library against your antigen, perform ELISA on them in E.coli, sequence the selected clones out of this, then cloning, transfection, and confirmation tests are completed before at least 5 positive clone sequences are sent via email.
Prellis – Their EXIS platform uses healthy donor B cells to make organoids which they then immunize with the antigen of interest. They can then get the antibody of interest from this immunized organoid. It boasts that it can reduce discovery time from 6-12 months in animals, to merely 3 weeks with EXIS, and also reduces costs.
Specifica – They use next-generation sequencing to produce antibody libraries in yeast or phage formats, as scFv or Fab libraries or common or variable light chains. Through the use of AI machine learning, they can split the libraries into clonotypes providing a whole host of antibodies against a variety of epitopes. They do initial screening with the Generation 3 phage display library (has natural CDR sequences, reducing liabilities and ensuring correct folding) then they use the yeast displayed library to narrow it down to fully binding antibodies. Each library they produce is unique to help avoid competitors choosing and developing the same therapeutic.
Tresars – Through Tresars use of display technology they provide fully reproducible and recombinant antibodies with serum-free methods, endeavouring to overcome the antibody reproducibility crisis without the use of animals.
YUMAB – YUMAB provides individually designed contract research projects for the discovery and the development of fully human antibodies, as well as R&D services for the engineering of antibodies.
10% discount on your next human biospecimen order from Tissue For Research (Accio BioBank Online) when you forward proof of purchase, or in-house generation, of animal-free recombinant antibodies. Additional discounts are available when ordering from our partners- please get in touch at info@biobankonline.com for more details of how you can benefit from this.
Information provided courtesy of companies’ websites. Accio Biobank Online is not responsible for any information about, or services provided by, external companies.
Abcalis – By using our sequence defined recombinant antibodies, you can obtain superior quality of results with animal-free reagents. It is our mission and aspiration every single day to achieve this goal, both for a sustainable future and medical safety of this and every future generation to come.
Abcam – They have a selection of animal-free antibodies and proteins generated in E.coli, yeast, synthetic, human embryonic kidney (HEK), or Chinese hamster ovary (CHO) cell expression systems ranging from fragments to full-length proteins. They no longer use the ascites method but instead use tissue culture and expression vectors. The antibodies they provide are for research purposes only but they can also be used commercially through a partnership with Abcam.
Absolute Antibody – Absolute Antibody provides Antibody Sequencing, Engineering & Recombinant Expression.
Adimab – Discovery of antibodies is via a yeast system with either synthetic or natural libraries incorporated. This system allows the selection of properties in real-time. They can also use phage display libraries and can optimize almost any property. They also offer the ability to create bispecifics based on IgG, scFv, and CD3.
AdipoGen Life Sciences – Through the use of human naïve phage display libraries they produce animal-free monoclonal antibodies. Their antibodies focus on functional activation/blocking of their targets and they also employ a selection strategy that allows the selection of antibodies based on custom functions such as targeting a specific conformational state antigen or identification of multi-protein complexes. They are available via AdipoGen’s custom services for research, pre-clinical and therapeutic applications.
Aptagen – Aptagen is a leading world-wide biotechnology company offering aptamer (synthetic antibody) products and services as research reagents, diagnostic and biomarker discovery tools, as well as for use in drug discovery and targeted delivery for therapeutics, and bioindustrial applications. We have a combined 25+ years of experience in creating aptamers that bind a wide variety of targets against small molecules, proteins, biomarkers and cells. Aptagen has performed this service for more than 50 companies, organizations, and institutions worldwide.
AvantGen – AvantGen has constructed 30 human antibody libraries (Germliner™) with multiple human antibody germline frameworks. These were selected based on their high stability, solubility and expression level. Diversity is focused in the CDR regions and mimics the diversity of the natural human antibody repertoire for that variable region family. The framework sequences are left as fully human germline sequences. AvantGen’s proprietary libraries are comprised of >100 billion members, are well expressed and displayed with AvantGen’s yeast display system allowing efficient human antibody discovery.
BioRad – These have produced thousands of completely animal-free antibodies. They mainly use phage display with a fully synthetic, naïve human combinatorial antibody library. Recently they have employed a new technique of antibody production called SpyTag that makes antibodies by protein ligation and site-directed conjugation within 8 weeks. With each antibody webpage, they have included a list of applications tested in-house and through information in peer-reviewed journals.
Cell Signalling Technology (CST) – They also dropped the ascites method in favor of cDNA and phage libraries expressed in vectors such as CHO cells, HEK cells, Pichia pastoris, etc. CST only provides recombinant antibodies for research and development use.
Geneva Antibody Facility – Providing recombinant antibodies to the academic community, not-for-profit. It is also known as the ABCD database, each antibody within this database is linked to the UniProt website for more information as well as its own ABCD detailed summary which includes all known research applications, this is constantly being updated as new information is published. It is also linked to a manufacturing company that can make these antibodies on demand. It consists of over 23,000 antibodies and 4000 antigens.
GenScript – GenScript offers one non-animal expression method using bacterial vectors and a variety of recombinant antibodies for research use. Their BacPower system uses E.coli as the expression vector and can produce proteins in an average of 4-6 weeks. They also offer another alternative to animal use in their insect expression system. This system infects baculovirus-infected cells with the recombinant plasmid to produce either virus or recombinant antibodies.
Human Antibody Core Facility – One of the few laboratories in the world that produces fully-human, full-length, antigen-specific antibodies for use in studying human immune responses. The Core has achieved breakthroughs in antibody technology and produced hundreds of high affinity protective antibodies to influenza, anthrax lethal toxin, and various S. pneumonia polysaccharides. The Core aims to support investigators by helping quantify the antibody secreting cell responses after vaccination and by generating human monoclonal antibodies to be characterized. Pathogen-specific human monoclonal antibodies are also available for licensing agreements and other forms of commercial development.
HYBRiBODY – They produce antibodies through phage display offering selection of recombinant antibodies and nanobodies, that are ELISA validated, for basic research and diagnostics. They also offer additional services to test the validated antibodies including cell imaging and can create ‘minibodies’ with the species-specific Fc region you require. They can employ Yeast two Hybrid technology after successive rounds of phage display to screen for intrabodies, which are antibodies that work inside living cells. Intrabodies are very useful for live cell imaging and in vivo applications.
ProMab – They provide a library of human Fab fragments only, isolated from 120 donors. These antibodies are present in their general recombinant and natural catalogs but more specific antibodies can be obtained from their custom services too. They pan the library against your antigen, perform ELISA on them in E.coli, sequence the selected clones out of this, then cloning, transfection, and confirmation tests are completed before at least 5 positive clone sequences are sent via email.
Prellis – Their EXIS platform uses healthy donor B cells to make organoids which they then immunize with the antigen of interest. They can then get the antibody of interest from this immunized organoid. It boasts that it can reduce discovery time from 6-12 months in animals, to merely 3 weeks with EXIS, and also reduces costs.
Specifica – They use next-generation sequencing to produce antibody libraries in yeast or phage formats, as scFv or Fab libraries or common or variable light chains. Through the use of AI machine learning, they can split the libraries into clonotypes providing a whole host of antibodies against a variety of epitopes. They do initial screening with the Generation 3 phage display library (has natural CDR sequences, reducing liabilities and ensuring correct folding) then they use the yeast displayed library to narrow it down to fully binding antibodies. Each library they produce is unique to help avoid competitors choosing and developing the same therapeutic.
Tresars – Through Tresars use of display technology they provide fully reproducible and recombinant antibodies with serum-free methods, endeavouring to overcome the antibody reproducibility crisis without the use of animals.
YUMAB – YUMAB provides individually designed contract research projects for the discovery and the development of fully human antibodies, as well as R&D services for the engineering of antibodies.
10% discount on your next human biospecimen order from Tissue For Research (Accio BioBank Online) when you forward proof of purchase, or in-house generation, of animal-free recombinant antibodies. Additional discounts are available when ordering from our partners- please get in touch at info@biobankonline.com for more details of how you can benefit from this.
9 Abbreviations
ADC – Antibody Drug Conjugate
ADCC – Antibody-Dependent Cellular Cytotoxic
ADCP – Antibody-Dependent Cellular Phagocytosis
AFA – Animal-Free Antibody
AI – Artificial Intelligence
ART-Ig – Asymmetric Re-engineering Technology – Immunoglobulin
ATD – Amino Terminal Domain
ATG16L – Autophagy Related 16 Like
1C – Constant
C1q – complement Component
1qCDC – Complement-Dependent Cytotoxicity
CDR – Complementarity Determining Region
CD – Cluster of Differentiation
CHO – Chinese Hamster Ovary cells
CNN – Convolutional Neural Network
CRISPR – Clustered Regularly Interspaced Short Palindromic Repeats
CST – Cell Signalling Technologies
E – Envelope protein
EBV – EBola Virus
EGFR – Epidermal Growth Factor Receptor
ELISA – Enzyme-Linked Immunosorbent Assay
ESAC – European Union Reference Laboratory for alternatives to animal testing (EURL ECVAM) Safety Advisory
Fab – Fragment antigen-binding
FACS – Fluorescent Activated Cell Sorting
Fc – Fragment crystallizable
FcεR – Fc epsilon Receptor
FDA – US Food and Drug Administration
Fv – Variable Fragment
GFP – Green Fluorescent Protein
GI – GastroIntestinalH – Heavy
HAMA – Human Anti-Mouse Antibody
HDR – Homology Directed Repair
HEK – Human Embryonic Kidney cells
HIV – Human Immunodeficiency Virus
HTS – High Throughput Screening
Ig – Immunoglobulin
IHC – ImmunoHistoChemical
INN – International Nonproprietary Names
L – Light
LGI1 – Leucine-rich glioma-inactivated
1LPS – LipoPolySaccharide
LSTM – Long Short Term Memory
mAb – monoclonal Antibody
MACS – Magnetic- Activated Cell Sorting
NGS – Next Generation Sequencing
NMDA – N-methyl-D-aspartate
NSCLC – Non-Small Cell Lung Cancer
OECD – the Organisation for Economic Co-operation and Development
pAb – polyclonal Antibody
PCR – Polymerase Chain Reaction
PETA – People for Ethical Treatment of AnimalsPPS – Pneumococcal polysaccharides
PRM – Protein-Ribosome-mRNA
PTM – Post Translational Modification
rAb – recombinant Antibody
RT-PCR – Reverse Transcriptase Polymerase Chain Reactions
cFv – single-chain Variable Fragment
TNF⍺ – Tumor Necrosis Factor-alpha
US NIH – US National Institutes of Health
V – Variable
ZIKV – ZIKa Virus
ADCC – Antibody-Dependent Cellular Cytotoxic
ADCP – Antibody-Dependent Cellular Phagocytosis
AFA – Animal-Free Antibody
AI – Artificial Intelligence
ART-Ig – Asymmetric Re-engineering Technology – Immunoglobulin
ATD – Amino Terminal Domain
ATG16L – Autophagy Related 16 Like
1C – Constant
C1q – complement Component
1qCDC – Complement-Dependent Cytotoxicity
CDR – Complementarity Determining Region
CD – Cluster of Differentiation
CHO – Chinese Hamster Ovary cells
CNN – Convolutional Neural Network
CRISPR – Clustered Regularly Interspaced Short Palindromic Repeats
CST – Cell Signalling Technologies
E – Envelope protein
EBV – EBola Virus
EGFR – Epidermal Growth Factor Receptor
ELISA – Enzyme-Linked Immunosorbent Assay
ESAC – European Union Reference Laboratory for alternatives to animal testing (EURL ECVAM) Safety Advisory
Fab – Fragment antigen-binding
FACS – Fluorescent Activated Cell Sorting
Fc – Fragment crystallizable
FcεR – Fc epsilon Receptor
FDA – US Food and Drug Administration
Fv – Variable Fragment
GFP – Green Fluorescent Protein
GI – GastroIntestinalH – Heavy
HAMA – Human Anti-Mouse Antibody
HDR – Homology Directed Repair
HEK – Human Embryonic Kidney cells
HIV – Human Immunodeficiency Virus
HTS – High Throughput Screening
Ig – Immunoglobulin
IHC – ImmunoHistoChemical
INN – International Nonproprietary Names
L – Light
LGI1 – Leucine-rich glioma-inactivated
1LPS – LipoPolySaccharide
LSTM – Long Short Term Memory
mAb – monoclonal Antibody
MACS – Magnetic- Activated Cell Sorting
NGS – Next Generation Sequencing
NMDA – N-methyl-D-aspartate
NSCLC – Non-Small Cell Lung Cancer
OECD – the Organisation for Economic Co-operation and Development
pAb – polyclonal Antibody
PCR – Polymerase Chain Reaction
PETA – People for Ethical Treatment of AnimalsPPS – Pneumococcal polysaccharides
PRM – Protein-Ribosome-mRNA
PTM – Post Translational Modification
rAb – recombinant Antibody
RT-PCR – Reverse Transcriptase Polymerase Chain Reactions
cFv – single-chain Variable Fragment
TNF⍺ – Tumor Necrosis Factor-alpha
US NIH – US National Institutes of Health
V – Variable
ZIKV – ZIKa Virus
10 References
1. Schroeder HW, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol 2010; 125: S41.
2. Kaufmann SHE. Immunology’s coming of age. Frontiers in Immunology 2019; 10: 684.
3. Nelson AL. Antibody fragments: Hope and hype. MAbs 2010; 2: 77.
4. Antibody purification application page, https://www.biotage.com/antibody-purification (accessed 15 July 2021).
5. Chiu ML, Goulet DR, Teplyakov A, et al.
6. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies 2019; 8: 55.
7. Antibody Structure | BioAtla, https://www.bioatla.com/appendix/antibody-structure/ (accessed 15 July 2021).
8. Ma H, O’Kennedy R. The structure of natural and recombinant antibodies. In: Methods in Molecular Biology. Humana Press Inc., pp. 7–11.Types of antibodies | MBL Life Science -JAPAN-, https://ruo.mbl.co.jp/bio/e/support/method/antibody-isotype.html (accessed 8 June 2021).
9. Human immunoglobulins. Transfusion Medicine and Hemotherapy 2009; 36: 449–459.
10. Complement fixation. In: Rheumatology and Immunology Therapy. Berlin/Heidelberg: Springer-Verlag, pp. 242–242.
11. Zuo Y, Evangelista F, Culton D, et al. IgG4 autoantibodies are inhibitory in the autoimmune disease bullous pemphigoid HHS Public Access. J Autoimmun 2016; 73: 111–119.
12. Elkon K, Casali P. Nature and functions of autoantibodies. Nature Clinical Practice Rheumatology 2008; 4: 491–498.
13. Chen K, Cerutti A. The function and regulation of immunoglobulin D. Current Opinion in Immunology 2011; 23: 345–352
14 .Woof JM, Ken MA. The function of immunoglobulin A in immunity. Journal of Pathology 2006; 208: 270–282.
15. Charles A Janeway J, Travers P, Walport M, et al. Effector mechanisms in allergic reactions, https://www.ncbi.nlm.nih.gov/books/NBK27112/ (2001, accessed 8 June 2021).
16. Sutton B, Davies A, Bax H, et al. IgE Antibodies: From Structure to Function and Clinical Translation. Antibodies 2019; 8: 19.
17. Antibody Types: IgM, IgA, IgD, IgG, IgE and Camelid Antibodies, https://www.news-medical.net/life-sciences/Types-of-Antibodies.aspx (accessed 15 July 2021).
18. Alberts B, Johnson A, Lewis J, et al. The Adaptive Immune System, https://www.ncbi.nlm.nih.gov/books/NBK21070/ (2002, accessed 8 June 2021).
19. Wang Q, Chen Y, Park J, et al. Design and Production of Bispecific Antibodies. Antibodies 2019; 8: 43.
20. Polyclonal Vs Monoclonal Vs Recombinant Antibodies: What’s the Difference?, https://blog.benchsci.com/polyclonal-vs-monoclonal-vs-recombinant-antibodies-whats-the-difference (accessed 8 June 2021)
21. .Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs 2017; 9: 182.
22. Johnson M. Antibody Structure and Fragments. Mater Methods; 3. Epub ahead of print 21 December 2013. DOI: 10.13070/MM.EN.3.160.
23. Antibodies and In Vivo Therapy. In: Immunology for Pharmacy. Elsevier, 2012, pp. 102–109.
24. Concepts in Immunology and Immunotherapeutics – Google Books, https://books.google.ie/books?hl=en&lr=&id=G46DrdlxNJAC&oi=fnd&pg=PR1&dq=Smith+BT:+Concepts+in+immunology+and+immunotherapeutics&ots=IOQo0drVL8&sig=r06gALBYONh88YJ8USu5n-o6Smk&redir_esc=y#v=onepage&q&f=false (accessed 15 July 2021).
25. Antibody Drug Nomenclature | BioAtla, https://www.bioatla.com/appendix/antibody-nomenclature/ (accessed 15 July 2021).
26. Groff K, Brown J, Clippinger AJ. Modern affinity reagents: Recombinant antibodies and aptamers. Biotechnol Adv 2015; 33: 1787–1798.
27. Marx V. Change-makers bring on recombinant antibodies. Nature Methods 2020; 17: 763–766.
28. Recombinant Antibodies: An Overview, https://info.gbiosciences.com/blog/recombinant-antibodies-an-overview (accessed 8 June 2021).
29. Reasons for Using Recombinant Antibodies – PETA Science Consortium International e.V., https://www.thepsci.eu/reasons-for-animal-free-antibodies/ (accessed 20 August 2021).
30. Gray A, Bradbury ARM, Knappik A, et al. Animal-free alternatives and the antibody iceberg. Nature Biotechnology 2020; 38: 1234–1239.
31. Better antibodies without using animals | EU Science Hub, https://ec.europa.eu/jrc/en/science-update/better-antibodies-without-using-animals (accessed 9 June 2021).
32. Joshi S, Yu D. Immunofluorescence. Basic Sci Methods Clin Res 2017; 135–150.
33. Janardhan KS, Jensen H, Clayton NP, et al. Immunohistochemistry in Investigative and Toxicologic Pathology. Toxicologic Pathology 2018; 46: 488–510.
34. The history of ELISA from creation to COVID-19 research | Molecular Devices, https://www.moleculardevices.com/lab-notes/microplate-readers/the-history-of-elisa#gref (accessed 8 June 2021).
35. Alhajj M, Farhana A. Enzyme Linked Immunosorbent Assay. StatPearls, https://www.ncbi.nlm.nih.gov/books/NBK555922/ (2021, accessed 20 August 2021).
36. Overview of Western Blotting | Thermo Fisher Scientific – IE, https://www.thermofisher.com/ie/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-western-blotting.html (accessed 8 June 2021).
37. Mahmood T, Yang P-C. Western Blot: Technique, Theory, and Trouble Shooting. N Am J Med Sci 2012; 4: 429.
38. A Brief History of Flow Cytometry | FLOW CONTRACT SITE, https://fcslaboratory.com/a-brief-history-of-flow-cytometry/ (accessed 8 June 2021).
39. McKinnon KM. Flow Cytometry: An Overview. Curr Protoc Immunol 2018; 120: 5.1.1.
40. Fukata M, Yokoi N, Fukata Y. Neurobiology of autoimmune encephalitis. Curr Opin Neurobiol 2018; 48: 1–8.
41. W T, R A, Z G, et al. An antibody for analysis of autophagy induction. Nat Methods 2020; 17: 232–239.
42. R M, NG S, R A, et al. Effect of maternal nutritional status on immunological substances in human colostrum and milk. Am J Clin Nutr 1983; 37: 632–640.
43. Sánchez-Salguero ES, Rodríguez-Chacón BC, Leyva-Daniel J, et al. ANTIGENIC STIMULATION DURING PREGNANCY MODIFIES SPECIFIC IGA1 AND IGA2 SUBCLASSES IN HUMAN COLOSTRUM ACCORDING TO THE CHEMICAL COMPOSITION OF THE ANTIGEN. Rev Invest Clin 2020; 72: 80–87.
44. Metz SW, Thomas A, Brackbill A, et al. Oligomeric state of the ZIKV E protein defines protective immune responses. Nat Commun 2019 101 2019; 10: 1–7.
45. B cells and food allergy : Current Opinion in Pediatrics, https://journals.lww.com/co-pediatrics/Abstract/9000/B_cells_and_food_allergy.98639.aspx (accessed 20 August 2021).
46. Nosanchuk JD. The interdependence of antibody C and V regions on specificity and affinity: Significant implications for the engineering of therapeutic antibodies. Virulence 2013; 4: 439.
47. Castelli MS, McGonigle P, Hornby PJ. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacology research & perspectives 2019; 7: e00535.
48. Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. mAbs 2015; 7: 9–14.
49. Mullard A. FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov. Epub ahead of print 5 May 2021. DOI: 10.1038/d41573-021-00079-7.
50. Foster RH, Wiseman LR. Abciximab. An updated review of its use in ischaemic heart disease. Drugs 1998; 56: 629–665.
51. Maloney DG, Grillo-López AJ, White CA, et al. IDEC-C2B8 (Rituximab) Anti-CD20 Monoclonal Antibody Therapy in Patients With Relapsed Low-Grade Non-Hodgkin’s Lymphoma. Blood 1997; 90: 2188–2195.
52. Almagro JC, Fransson J. Humanization of antibodies. Frontiers in Bioscience 2008; 13: 1619–1633.
53. Uchiyama T, Broder S, Waldmann TA. A monoclonal antibody (anti-Tac) reactive with activated and functionally mature human T cells. I. Production of anti-Tac monoclonal antibody and distribution of Tac (+) cells. J Immunol 1981; 126: 1393–137.
54. Brüggemann M, Osborn MJ, Ma B, et al. Human Antibody Production in Transgenic Animals. Archivum Immunologiae et Therapiae Experimentalis 2015; 63: 101–108.
55. Gibson TB, Ranganathan A, Grothey A. Randomized phase III trial results of Panitumumab, a fully human anti-epidermal growth factor receptor monoclonal antibody, in metastatic colorectal cancer. Clinical Colorectal Cancer 2006; 6: 29–31.
56. Jain M, Kamal N, Batra SK. Engineering antibodies for clinical applications. Trends Biotechnol 2007; 25: 307–316.
57. McCafferty J, Griffiths AD, Winter G, et al. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 1990; 348: 552–554.
58. The Nobel Prize in Chemistry 2018 – NobelPrize.org, https://www.nobelprize.org/prizes/chemistry/2018/summary/ (accessed 8 June 2021).
59. Reader RH, Workman RG, Maddison BC, et al. Advances in the Production and Batch Reformatting of Phage Antibody Libraries. Molecular Biotechnology 2019; 61: 801–815.
60. L L, M K, A K-V, et al. Basics of Antibody Phage Display Technology. Toxins (Basel); 10. Epub ahead of print 9 June 2018. DOI: 10.3390/TOXINS10060236.
61. Smaglo BG, Aldeghaither D, Weiner LM. Antibody Therapy. Encycl Immunobiol 2016; 4: 550–559.
62. Chao G, Lau WL, Hackel BJ, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc 2006 12 2006; 1: 755–768.
63. Frenzel A, Kügler J, Helmsing S, et al. Designing Human Antibodies by Phage Display. Transfusion Medicine and Hemotherapy 2017; 44: 312–318.
64. Phage Display and Antibody Library | Sino Biological, https://www.sinobiological.com/resource/antibody-technical/phage-display-antibody (accessed 20 August 2021).
65. Recombinant antibodies: infinitely better | Abcam, https://www.abcam.com/primary-antibodies/recombinant-antibodies (accessed 8 June 2021).
66. Kempeni J. Preliminary results of early clinical trials with the fully human anti- TNFα monoclonal antibody D2E7. Annals of the Rheumatic Diseases 1999; 58: I70.
67. A B, A Z, M H, et al. Single Chain Antibodies as Tools to Study transforming growth factor-β-Regulated SMAD Proteins in Proximity Ligation-Based Pharmacological Screens. Mol Cell Proteomics 2016; 15: 1848–1856.
68. Alfaleh MA, Alsaab HO, Mahmoud AB, et al. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Frontiers in Immunology 2020; 11: 1986.
69. Mattheakis LC, Bhatt RR, Dower WJ. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci 1994; 91: 9022–9026.
70. Osbourn JK. Biological Macromolecules. In: Comprehensive Medicinal Chemistry II. Elsevier, pp. 431–447.
71. Kunamneni A, Ogaugwu C, Bradfute S, et al. Ribosome Display Technology: Applications in Disease Diagnosis and Control. Antibodies 2020; 9: 28.
72. Feldhaus MJ, Siegel RW. Yeast display of antibody fragments: a discovery and characterization platform. J Immunol Methods 2004; 290: 69–80.
73. Uchański T, Zögg T, Yin J, et al. An improved yeast surface display platform for the screening of nanobody immune libraries. Sci Reports 2019 91 2019; 9: 1–12.
74. Surface Technology FAQ – Creative Biolabs, https://www.creative-biolabs.com/surface-technology-faq.html (accessed 9 June 2021).
75. Marks JD. Monoclonal Antibodies from Display Libraries. In: Molecular Biology of B Cells. Elsevier Inc., 2004, pp. 511–531.
76. Bradbury ARM, Sidhu S, Dübel S, et al. Beyond natural antibodies: The power of in vitro display technologies. Nature Biotechnology 2011; 29: 245–254.
77. Tiller T, Meffre E, Yurasov S, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008; 329: 112–124.
78. Tiller T. Single B cell antibody technologies. New Biotechnology 2011; 28: 453–457.
79. Ojima-Kato T, Morishita S, Uchida Y, et al. Rapid Generation of Monoclonal Antibodies from Single B Cells by Ecobody Technology. Antibodies 2018; 7: 38.
80. Rudkin FM, Raziunaite I, Workman H, et al. Single human B cell-derived monoclonal anti-Candida antibodies enhance phagocytosis and protect against disseminated candidiasis. Nat Commun 2018; 9: 1–16.
81. Wycoff K. Secretory IgA Antibodies from Plants. Curr Pharm Des 2005; 11: 2429–2437.
82. Jain P, Pandey P, Jain D, et al. PLANTIBODY: AN OVERVIEW. undefined.
83. AS M. Exploring transgenic plants as a new vaccine source. Science 1995; 268: 658–660.
84. A H, R C, K B. Production of antibodies in transgenic plants. Nature 1989; 342: 76–78.
85. Oluwayelu DO, Adebiyi AI. Plantibodies in human and animal health: a review. Afr Health Sci 2016; 16: 640.
86. U F, U C. High-level production and long-term storage of engineered antibodies in transgenic tobacco seeds. Biotechnology (N Y) 1995; 13: 1090–1093.
87. Ghag SB, Adki VS, Ganapathi TR, et al. Plant Platforms for Efficient Heterologous Protein Production. Biotechnol Bioprocess Eng 2021; 26: 1.
88. NOPR: Transgenic plants as bioreactors, http://nopr.niscair.res.in/handle/123456789/5856 (accessed 14 September 2021).
89. K H, U S. Production of new/modified proteins in transgenic plants. Curr Opin Biotechnol 1999; 10: 163–168.
90. R F, RM T, S S. Production of antibodies in plants and their use for global health. Vaccine 2003; 21: 820–825.
91. Icon Genetics, https://www.icongenetics.com/ (accessed 14 September 2021).
92. Plantibodies Could Pave the Road to Wellness, https://www.newsweek.com/2014/12/26/plantibodies-could-pave-road-wellness-291950.html (accessed 14 September 2021).
93. Liao Y-C, Pingli H, Senzhao C, et al. Biotechnology and Genetic Engineering Reviews Plantibodies: A Novel Strategy to Create Pathogen-Resistant Plants 13 Plantibodies: A Novel Strategy to Create Pathogen-Resistant Plants. DOI: 10.1080/02648725.2006.10648087.
94. Frenzel A, Hust M, Schirrmann T. Expression of Recombinant Antibodies. Front Immunol 2013; 0: 217.
95. New Presentation on Animal-Free Recombinant Antibodies, https://absoluteantibody.com/general/new-presentation-on-animal-free-recombinant-antibodies/ (accessed 9 June 2021).
96. Gray AC, Bradbury ARM, Knappik A, et al. Animal-derived-antibody generation faces strict reform in accordance with European Union policy on animal use. Nature Methods 2020; 17: 755–756.
97. Jensen MM, Karring H. The origins and developments of sulfation-prone tyrosine-rich and acidic N- and C-terminal extensions of class ll and lll small leucine-rich repeat proteins shed light on connective tissue evolution in vertebrates. BMC Evol Biol 2020; 20: 1–17.
98. Complete Guide to Recombinant Antibodies | Benefits & Uses, https://www.activemotif.com/blog-recombinant-abs (accessed 20 August 2021).
99. EUR-Lex – 32010L0063 – EN – EUR-Lex, https://eur-lex.europa.eu/eli/dir/2010/63/oj (accessed 9 June 2021).
100. The case for replacement Afability Animal Friendly Affinity Reagents, http://www.afability.com/services-view/the-case-for-replacement/ (accessed 9 June 2021).
101. JRC Publications Repository – EURL ECVAM Recommendation on Non-Animal-Derived Antibodies, https://publications.jrc.ec.europa.eu/repository/handle/JRC120199 (accessed 9 June 2021).
102. Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nat 2015 5187537 2015; 518: 27–29.
103. Saka K, Kakuzaki T, Metsugi S, et al. Antibody design using LSTM based deep generative model from phage display library for affinity maturation. Sci Reports 2021 111 2021; 11: 1–13.
104. Y X, QH L, F H, et al. An efficient gene selection method for microarray data based on LASSO and BPSO. BMC Bioinformatics; 20. Epub ahead of print 30 December 2019. DOI: 10.1186/S12859-019-3228-0.
105. Mason DM, Friedensohn S, Weber CR, et al. Deep learning enables therapeutic antibody optimization in mammalian cells by deciphering high-dimensional protein sequence space. bioRxiv 2019; 617860.
106. Laustsen AH, Greiff V, Karatt-Vellatt A, et al. Animal Immunization, in Vitro Display Technologies, and Machine Learning for Antibody Discovery. Trends Biotechnol; 0. Epub ahead of print 2021. DOI: 10.1016/J.TIBTECH.2021.03.003.
107. Prabakaran P, Rao SP, Wendt M. Animal immunization merges with innovative technologies: A new paradigm shift in antibody discovery. MAbs; 13. Epub ahead of print 2021. DOI: 10.1080/19420862.2021.1924347.
108. Thakur A, Huang M, Lum LG. Bispecific antibody based therapeutics: Strengths and challenges. Blood Rev 2018; 32: 339–347.
109. Shim H. Bispecific Antibodies and Antibody–Drug Conjugates for Cancer Therapy: Technological Considerations. Biomolecules; 10. Epub ahead of print 1 March 2020. DOI: 10.3390/BIOM10030360.
110. A N, MM R. Recombination of a mixture of univalent antibody fragments of different specificity. Arch Biochem Biophys 1961; 93: 460–462.
111. Müller D, Kontermann RE. Bispecific Antibodies for Cancer Immunotherapy. BioDrugs 2010 242 2012; 24: 89–98.T I. [Next Generation 112. Antibody Therapeutics Using Bispecific Antibody Technology]. Yakugaku Zasshi 2017; 137: 831–836.
113. Neovii completes marketing authorisation withdrawal of Removab® in the European Union | Neovii, https://neovii.com/neovii-completes-marketing-authorisation-withdrawal-of-removab-in-the-european-union/ (accessed 15 July 2021).
114. FDA grants accelerated approval to amivantamab-vmjw for metastatic non-small cell lung cancer | FDA, https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-amivantamab-vmjw-metastatic-non-small-cell-lung-cancer (accessed 15 July 2021).
115. FDA approves emicizumab-kxwh for hemophilia A with or without factor VIII inhibitors | FDA, https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-emicizumab-kxwh-hemophilia-or-without-factor-viii-inhibitors (accessed 15 July 2021).
116. FDA modifies immunogenicity language in final bispecific antibody guidance | RAPS, https://www.raps.org/news-and-articles/news-articles/2021/5/antibody (accessed 15 July 2021).
2. Kaufmann SHE. Immunology’s coming of age. Frontiers in Immunology 2019; 10: 684.
3. Nelson AL. Antibody fragments: Hope and hype. MAbs 2010; 2: 77.
4. Antibody purification application page, https://www.biotage.com/antibody-purification (accessed 15 July 2021).
5. Chiu ML, Goulet DR, Teplyakov A, et al.
6. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies 2019; 8: 55.
7. Antibody Structure | BioAtla, https://www.bioatla.com/appendix/antibody-structure/ (accessed 15 July 2021).
8. Ma H, O’Kennedy R. The structure of natural and recombinant antibodies. In: Methods in Molecular Biology. Humana Press Inc., pp. 7–11.Types of antibodies | MBL Life Science -JAPAN-, https://ruo.mbl.co.jp/bio/e/support/method/antibody-isotype.html (accessed 8 June 2021).
9. Human immunoglobulins. Transfusion Medicine and Hemotherapy 2009; 36: 449–459.
10. Complement fixation. In: Rheumatology and Immunology Therapy. Berlin/Heidelberg: Springer-Verlag, pp. 242–242.
11. Zuo Y, Evangelista F, Culton D, et al. IgG4 autoantibodies are inhibitory in the autoimmune disease bullous pemphigoid HHS Public Access. J Autoimmun 2016; 73: 111–119.
12. Elkon K, Casali P. Nature and functions of autoantibodies. Nature Clinical Practice Rheumatology 2008; 4: 491–498.
13. Chen K, Cerutti A. The function and regulation of immunoglobulin D. Current Opinion in Immunology 2011; 23: 345–352
14 .Woof JM, Ken MA. The function of immunoglobulin A in immunity. Journal of Pathology 2006; 208: 270–282.
15. Charles A Janeway J, Travers P, Walport M, et al. Effector mechanisms in allergic reactions, https://www.ncbi.nlm.nih.gov/books/NBK27112/ (2001, accessed 8 June 2021).
16. Sutton B, Davies A, Bax H, et al. IgE Antibodies: From Structure to Function and Clinical Translation. Antibodies 2019; 8: 19.
17. Antibody Types: IgM, IgA, IgD, IgG, IgE and Camelid Antibodies, https://www.news-medical.net/life-sciences/Types-of-Antibodies.aspx (accessed 15 July 2021).
18. Alberts B, Johnson A, Lewis J, et al. The Adaptive Immune System, https://www.ncbi.nlm.nih.gov/books/NBK21070/ (2002, accessed 8 June 2021).
19. Wang Q, Chen Y, Park J, et al. Design and Production of Bispecific Antibodies. Antibodies 2019; 8: 43.
20. Polyclonal Vs Monoclonal Vs Recombinant Antibodies: What’s the Difference?, https://blog.benchsci.com/polyclonal-vs-monoclonal-vs-recombinant-antibodies-whats-the-difference (accessed 8 June 2021)
21. .Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs 2017; 9: 182.
22. Johnson M. Antibody Structure and Fragments. Mater Methods; 3. Epub ahead of print 21 December 2013. DOI: 10.13070/MM.EN.3.160.
23. Antibodies and In Vivo Therapy. In: Immunology for Pharmacy. Elsevier, 2012, pp. 102–109.
24. Concepts in Immunology and Immunotherapeutics – Google Books, https://books.google.ie/books?hl=en&lr=&id=G46DrdlxNJAC&oi=fnd&pg=PR1&dq=Smith+BT:+Concepts+in+immunology+and+immunotherapeutics&ots=IOQo0drVL8&sig=r06gALBYONh88YJ8USu5n-o6Smk&redir_esc=y#v=onepage&q&f=false (accessed 15 July 2021).
25. Antibody Drug Nomenclature | BioAtla, https://www.bioatla.com/appendix/antibody-nomenclature/ (accessed 15 July 2021).
26. Groff K, Brown J, Clippinger AJ. Modern affinity reagents: Recombinant antibodies and aptamers. Biotechnol Adv 2015; 33: 1787–1798.
27. Marx V. Change-makers bring on recombinant antibodies. Nature Methods 2020; 17: 763–766.
28. Recombinant Antibodies: An Overview, https://info.gbiosciences.com/blog/recombinant-antibodies-an-overview (accessed 8 June 2021).
29. Reasons for Using Recombinant Antibodies – PETA Science Consortium International e.V., https://www.thepsci.eu/reasons-for-animal-free-antibodies/ (accessed 20 August 2021).
30. Gray A, Bradbury ARM, Knappik A, et al. Animal-free alternatives and the antibody iceberg. Nature Biotechnology 2020; 38: 1234–1239.
31. Better antibodies without using animals | EU Science Hub, https://ec.europa.eu/jrc/en/science-update/better-antibodies-without-using-animals (accessed 9 June 2021).
32. Joshi S, Yu D. Immunofluorescence. Basic Sci Methods Clin Res 2017; 135–150.
33. Janardhan KS, Jensen H, Clayton NP, et al. Immunohistochemistry in Investigative and Toxicologic Pathology. Toxicologic Pathology 2018; 46: 488–510.
34. The history of ELISA from creation to COVID-19 research | Molecular Devices, https://www.moleculardevices.com/lab-notes/microplate-readers/the-history-of-elisa#gref (accessed 8 June 2021).
35. Alhajj M, Farhana A. Enzyme Linked Immunosorbent Assay. StatPearls, https://www.ncbi.nlm.nih.gov/books/NBK555922/ (2021, accessed 20 August 2021).
36. Overview of Western Blotting | Thermo Fisher Scientific – IE, https://www.thermofisher.com/ie/en/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/overview-western-blotting.html (accessed 8 June 2021).
37. Mahmood T, Yang P-C. Western Blot: Technique, Theory, and Trouble Shooting. N Am J Med Sci 2012; 4: 429.
38. A Brief History of Flow Cytometry | FLOW CONTRACT SITE, https://fcslaboratory.com/a-brief-history-of-flow-cytometry/ (accessed 8 June 2021).
39. McKinnon KM. Flow Cytometry: An Overview. Curr Protoc Immunol 2018; 120: 5.1.1.
40. Fukata M, Yokoi N, Fukata Y. Neurobiology of autoimmune encephalitis. Curr Opin Neurobiol 2018; 48: 1–8.
41. W T, R A, Z G, et al. An antibody for analysis of autophagy induction. Nat Methods 2020; 17: 232–239.
42. R M, NG S, R A, et al. Effect of maternal nutritional status on immunological substances in human colostrum and milk. Am J Clin Nutr 1983; 37: 632–640.
43. Sánchez-Salguero ES, Rodríguez-Chacón BC, Leyva-Daniel J, et al. ANTIGENIC STIMULATION DURING PREGNANCY MODIFIES SPECIFIC IGA1 AND IGA2 SUBCLASSES IN HUMAN COLOSTRUM ACCORDING TO THE CHEMICAL COMPOSITION OF THE ANTIGEN. Rev Invest Clin 2020; 72: 80–87.
44. Metz SW, Thomas A, Brackbill A, et al. Oligomeric state of the ZIKV E protein defines protective immune responses. Nat Commun 2019 101 2019; 10: 1–7.
45. B cells and food allergy : Current Opinion in Pediatrics, https://journals.lww.com/co-pediatrics/Abstract/9000/B_cells_and_food_allergy.98639.aspx (accessed 20 August 2021).
46. Nosanchuk JD. The interdependence of antibody C and V regions on specificity and affinity: Significant implications for the engineering of therapeutic antibodies. Virulence 2013; 4: 439.
47. Castelli MS, McGonigle P, Hornby PJ. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacology research & perspectives 2019; 7: e00535.
48. Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. mAbs 2015; 7: 9–14.
49. Mullard A. FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov. Epub ahead of print 5 May 2021. DOI: 10.1038/d41573-021-00079-7.
50. Foster RH, Wiseman LR. Abciximab. An updated review of its use in ischaemic heart disease. Drugs 1998; 56: 629–665.
51. Maloney DG, Grillo-López AJ, White CA, et al. IDEC-C2B8 (Rituximab) Anti-CD20 Monoclonal Antibody Therapy in Patients With Relapsed Low-Grade Non-Hodgkin’s Lymphoma. Blood 1997; 90: 2188–2195.
52. Almagro JC, Fransson J. Humanization of antibodies. Frontiers in Bioscience 2008; 13: 1619–1633.
53. Uchiyama T, Broder S, Waldmann TA. A monoclonal antibody (anti-Tac) reactive with activated and functionally mature human T cells. I. Production of anti-Tac monoclonal antibody and distribution of Tac (+) cells. J Immunol 1981; 126: 1393–137.
54. Brüggemann M, Osborn MJ, Ma B, et al. Human Antibody Production in Transgenic Animals. Archivum Immunologiae et Therapiae Experimentalis 2015; 63: 101–108.
55. Gibson TB, Ranganathan A, Grothey A. Randomized phase III trial results of Panitumumab, a fully human anti-epidermal growth factor receptor monoclonal antibody, in metastatic colorectal cancer. Clinical Colorectal Cancer 2006; 6: 29–31.
56. Jain M, Kamal N, Batra SK. Engineering antibodies for clinical applications. Trends Biotechnol 2007; 25: 307–316.
57. McCafferty J, Griffiths AD, Winter G, et al. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 1990; 348: 552–554.
58. The Nobel Prize in Chemistry 2018 – NobelPrize.org, https://www.nobelprize.org/prizes/chemistry/2018/summary/ (accessed 8 June 2021).
59. Reader RH, Workman RG, Maddison BC, et al. Advances in the Production and Batch Reformatting of Phage Antibody Libraries. Molecular Biotechnology 2019; 61: 801–815.
60. L L, M K, A K-V, et al. Basics of Antibody Phage Display Technology. Toxins (Basel); 10. Epub ahead of print 9 June 2018. DOI: 10.3390/TOXINS10060236.
61. Smaglo BG, Aldeghaither D, Weiner LM. Antibody Therapy. Encycl Immunobiol 2016; 4: 550–559.
62. Chao G, Lau WL, Hackel BJ, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc 2006 12 2006; 1: 755–768.
63. Frenzel A, Kügler J, Helmsing S, et al. Designing Human Antibodies by Phage Display. Transfusion Medicine and Hemotherapy 2017; 44: 312–318.
64. Phage Display and Antibody Library | Sino Biological, https://www.sinobiological.com/resource/antibody-technical/phage-display-antibody (accessed 20 August 2021).
65. Recombinant antibodies: infinitely better | Abcam, https://www.abcam.com/primary-antibodies/recombinant-antibodies (accessed 8 June 2021).
66. Kempeni J. Preliminary results of early clinical trials with the fully human anti- TNFα monoclonal antibody D2E7. Annals of the Rheumatic Diseases 1999; 58: I70.
67. A B, A Z, M H, et al. Single Chain Antibodies as Tools to Study transforming growth factor-β-Regulated SMAD Proteins in Proximity Ligation-Based Pharmacological Screens. Mol Cell Proteomics 2016; 15: 1848–1856.
68. Alfaleh MA, Alsaab HO, Mahmoud AB, et al. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Frontiers in Immunology 2020; 11: 1986.
69. Mattheakis LC, Bhatt RR, Dower WJ. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci 1994; 91: 9022–9026.
70. Osbourn JK. Biological Macromolecules. In: Comprehensive Medicinal Chemistry II. Elsevier, pp. 431–447.
71. Kunamneni A, Ogaugwu C, Bradfute S, et al. Ribosome Display Technology: Applications in Disease Diagnosis and Control. Antibodies 2020; 9: 28.
72. Feldhaus MJ, Siegel RW. Yeast display of antibody fragments: a discovery and characterization platform. J Immunol Methods 2004; 290: 69–80.
73. Uchański T, Zögg T, Yin J, et al. An improved yeast surface display platform for the screening of nanobody immune libraries. Sci Reports 2019 91 2019; 9: 1–12.
74. Surface Technology FAQ – Creative Biolabs, https://www.creative-biolabs.com/surface-technology-faq.html (accessed 9 June 2021).
75. Marks JD. Monoclonal Antibodies from Display Libraries. In: Molecular Biology of B Cells. Elsevier Inc., 2004, pp. 511–531.
76. Bradbury ARM, Sidhu S, Dübel S, et al. Beyond natural antibodies: The power of in vitro display technologies. Nature Biotechnology 2011; 29: 245–254.
77. Tiller T, Meffre E, Yurasov S, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008; 329: 112–124.
78. Tiller T. Single B cell antibody technologies. New Biotechnology 2011; 28: 453–457.
79. Ojima-Kato T, Morishita S, Uchida Y, et al. Rapid Generation of Monoclonal Antibodies from Single B Cells by Ecobody Technology. Antibodies 2018; 7: 38.
80. Rudkin FM, Raziunaite I, Workman H, et al. Single human B cell-derived monoclonal anti-Candida antibodies enhance phagocytosis and protect against disseminated candidiasis. Nat Commun 2018; 9: 1–16.
81. Wycoff K. Secretory IgA Antibodies from Plants. Curr Pharm Des 2005; 11: 2429–2437.
82. Jain P, Pandey P, Jain D, et al. PLANTIBODY: AN OVERVIEW. undefined.
83. AS M. Exploring transgenic plants as a new vaccine source. Science 1995; 268: 658–660.
84. A H, R C, K B. Production of antibodies in transgenic plants. Nature 1989; 342: 76–78.
85. Oluwayelu DO, Adebiyi AI. Plantibodies in human and animal health: a review. Afr Health Sci 2016; 16: 640.
86. U F, U C. High-level production and long-term storage of engineered antibodies in transgenic tobacco seeds. Biotechnology (N Y) 1995; 13: 1090–1093.
87. Ghag SB, Adki VS, Ganapathi TR, et al. Plant Platforms for Efficient Heterologous Protein Production. Biotechnol Bioprocess Eng 2021; 26: 1.
88. NOPR: Transgenic plants as bioreactors, http://nopr.niscair.res.in/handle/123456789/5856 (accessed 14 September 2021).
89. K H, U S. Production of new/modified proteins in transgenic plants. Curr Opin Biotechnol 1999; 10: 163–168.
90. R F, RM T, S S. Production of antibodies in plants and their use for global health. Vaccine 2003; 21: 820–825.
91. Icon Genetics, https://www.icongenetics.com/ (accessed 14 September 2021).
92. Plantibodies Could Pave the Road to Wellness, https://www.newsweek.com/2014/12/26/plantibodies-could-pave-road-wellness-291950.html (accessed 14 September 2021).
93. Liao Y-C, Pingli H, Senzhao C, et al. Biotechnology and Genetic Engineering Reviews Plantibodies: A Novel Strategy to Create Pathogen-Resistant Plants 13 Plantibodies: A Novel Strategy to Create Pathogen-Resistant Plants. DOI: 10.1080/02648725.2006.10648087.
94. Frenzel A, Hust M, Schirrmann T. Expression of Recombinant Antibodies. Front Immunol 2013; 0: 217.
95. New Presentation on Animal-Free Recombinant Antibodies, https://absoluteantibody.com/general/new-presentation-on-animal-free-recombinant-antibodies/ (accessed 9 June 2021).
96. Gray AC, Bradbury ARM, Knappik A, et al. Animal-derived-antibody generation faces strict reform in accordance with European Union policy on animal use. Nature Methods 2020; 17: 755–756.
97. Jensen MM, Karring H. The origins and developments of sulfation-prone tyrosine-rich and acidic N- and C-terminal extensions of class ll and lll small leucine-rich repeat proteins shed light on connective tissue evolution in vertebrates. BMC Evol Biol 2020; 20: 1–17.
98. Complete Guide to Recombinant Antibodies | Benefits & Uses, https://www.activemotif.com/blog-recombinant-abs (accessed 20 August 2021).
99. EUR-Lex – 32010L0063 – EN – EUR-Lex, https://eur-lex.europa.eu/eli/dir/2010/63/oj (accessed 9 June 2021).
100. The case for replacement Afability Animal Friendly Affinity Reagents, http://www.afability.com/services-view/the-case-for-replacement/ (accessed 9 June 2021).
101. JRC Publications Repository – EURL ECVAM Recommendation on Non-Animal-Derived Antibodies, https://publications.jrc.ec.europa.eu/repository/handle/JRC120199 (accessed 9 June 2021).
102. Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nat 2015 5187537 2015; 518: 27–29.
103. Saka K, Kakuzaki T, Metsugi S, et al. Antibody design using LSTM based deep generative model from phage display library for affinity maturation. Sci Reports 2021 111 2021; 11: 1–13.
104. Y X, QH L, F H, et al. An efficient gene selection method for microarray data based on LASSO and BPSO. BMC Bioinformatics; 20. Epub ahead of print 30 December 2019. DOI: 10.1186/S12859-019-3228-0.
105. Mason DM, Friedensohn S, Weber CR, et al. Deep learning enables therapeutic antibody optimization in mammalian cells by deciphering high-dimensional protein sequence space. bioRxiv 2019; 617860.
106. Laustsen AH, Greiff V, Karatt-Vellatt A, et al. Animal Immunization, in Vitro Display Technologies, and Machine Learning for Antibody Discovery. Trends Biotechnol; 0. Epub ahead of print 2021. DOI: 10.1016/J.TIBTECH.2021.03.003.
107. Prabakaran P, Rao SP, Wendt M. Animal immunization merges with innovative technologies: A new paradigm shift in antibody discovery. MAbs; 13. Epub ahead of print 2021. DOI: 10.1080/19420862.2021.1924347.
108. Thakur A, Huang M, Lum LG. Bispecific antibody based therapeutics: Strengths and challenges. Blood Rev 2018; 32: 339–347.
109. Shim H. Bispecific Antibodies and Antibody–Drug Conjugates for Cancer Therapy: Technological Considerations. Biomolecules; 10. Epub ahead of print 1 March 2020. DOI: 10.3390/BIOM10030360.
110. A N, MM R. Recombination of a mixture of univalent antibody fragments of different specificity. Arch Biochem Biophys 1961; 93: 460–462.
111. Müller D, Kontermann RE. Bispecific Antibodies for Cancer Immunotherapy. BioDrugs 2010 242 2012; 24: 89–98.T I. [Next Generation 112. Antibody Therapeutics Using Bispecific Antibody Technology]. Yakugaku Zasshi 2017; 137: 831–836.
113. Neovii completes marketing authorisation withdrawal of Removab® in the European Union | Neovii, https://neovii.com/neovii-completes-marketing-authorisation-withdrawal-of-removab-in-the-european-union/ (accessed 15 July 2021).
114. FDA grants accelerated approval to amivantamab-vmjw for metastatic non-small cell lung cancer | FDA, https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-amivantamab-vmjw-metastatic-non-small-cell-lung-cancer (accessed 15 July 2021).
115. FDA approves emicizumab-kxwh for hemophilia A with or without factor VIII inhibitors | FDA, https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-emicizumab-kxwh-hemophilia-or-without-factor-viii-inhibitors (accessed 15 July 2021).
116. FDA modifies immunogenicity language in final bispecific antibody guidance | RAPS, https://www.raps.org/news-and-articles/news-articles/2021/5/antibody (accessed 15 July 2021).
